Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(7):533-539. doi:10.7150/ijms.14871 This issue Cite
Research Paper
Expression of SRSF3 is Correlated with Carcinogenesis and Progression of Oral Squamous Cell Carcinoma
Hubei-MOST KLOS & KLOBME, School & Hospital of Stomatology, Wuhan University, Wuhan, 430079, PR China.
*These authors contribute equally.
Received 2016-1-3; Accepted 2016-5-8; Published 2016-6-30
Abstract
Objective: Oral squamous cell carcinoma (OSCC) is the most common malignancy of head and neck with high mortality rates. The mechanisms of initiation and development of OSCC remain largely unknown. Dysregulated alternative splicing of pre-mRNA has been associated with OSCC. Splicing factor SRSF3 is a proto-oncogene and overexpressed in multiple cancers. The aim of this study was to uncover the relationship between SRSF3 and carcinogenesis and progression of oral squamous cell carcinoma.
Design and Methods: The expression of SRSF3 in oral normal, dysplasia, or carcinoma tissues was analyzed by immunohistochemistry. The expression levels of EMT-related genes were quantified by real-time quantitative RT-PCR. The expression of SRSF3 in DMBA treated primary cultured oral epithelial cells were analyzed by western blot.
Result: SRSF3 is overexpressed in oral cancer and moderate or severe dysplasia tissues. Patients with high grade cancer or lymphatic metastasis showed up-regulated expression of SRSF3. Knockdown of SRSF3 repressed the expression of Snail and N-cadherin in vitro. Carcinogen DMBA treated primary cultured oral epithelial cells showed significantly increased SRSF3 level than in control cells.
Conclusion: Our results suggested that SRSF3 is associated with the initiation and development of OSCC and may be a biomarker and therapeutic target of OSCC.
Keywords: SRSF3, oral squamous cell carcinoma, metastasis.
Introduction
Oral squamous cell carcinoma (OSCC) is the most common malignancy of head and neck. [1]. The mortality rates of OSCC are still high despite progress of treatment and diagnosis in the past decades [2]. It is urgently required to uncover the mechanisms of initiation and development of OSCC. It has been demonstrated that dysregulated alternative splicing of pre-mRNA is associated with OSCC [3, 4]. Several splicing factors have been shown to be involved in the tumorigenesis of OSCC [5, 6].
SR proteins (serine/arginine-rich proteins), functioning as messenger-RNA-binding proteins and alternative splicing factors, all contain one or two RNA recognition motif(s) (RRM) and a protein-interaction arginine-serine rich (RS) domain [7]. SRSF3 (serine/arginine-rich splicing factor 3), also known as SRp20 or SFRS3, is the smallest member of SR proteins [8]. SRSF3 is a multi-functional protein. It has been shown to be involved in multiple cellular functions, such as alternative splicing [9, 10], DNA repair [11], RNA export [12, 13], alternative RNA polyadenylation [14], and protein translation [15, 16]. SRSF3 has been found to be involved in a number of human diseases, including cancer [17]. It has been demonstrated to be a proto-oncogene and often overexpressed in multiple cancers, including OSCC [18, 19]. However, the relationship between SRSF3 and carcinogenesis and caner progression remains unclear.
In this study, we analyzed the expression of SRSF3 in oral precancerous and cancerous tissues. We found that SRSF3 is overexpressed in oral cancer and moderate or severe dysplasia tissues. Patients with high grade cancer or lymphatic metastasis showed up-regulated expression of SRSF3.
Materials and Methods
Cells and RNAi
FaDu cells (ATCC) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone, USA) and 1% antibiotic-antimycotic (Invitrogen, USA). SRSF3 siRNA#1 and #2 were purchased from Santa Cruz Biotechnology and Ambion, respectively. FaDu cells were transfected with 20nM siRNA in the presence of Lipofentamine 2000 (Invitrogen, USA) according to the manufacturer's instructions. After 48h, cells were passed and received another transfection. After 96h, total proteins were collected by adding 2× sodium dodecyl sulfate sample buffer, and boiled for 5 min. Total RNAs were purified by TRIzol reagent (Invitrogen, USA).
Patients and Tissue Samples
Seventy-seven patients diagnosed as oral squamous cell carcinoma and eleven patients diagnosed as epithelial dysplasia were involved in this study. All histologic diagnoses were performed by the pathology department in the School and Hospital of Stomatology, Wuhan University. Normal oral mucosal tissues were obtained from non-tumor adjacent tissues or patients with reshaping of gingival tissues. Informed consent was obtained from all subjects. All experimental protocols were approved by the Ethics Committee at the School and Hospital of Stomatology in Wuhan University.
Immunohistochemistry
Immunohistochemical staining was performed using mouse anti-SRSF3 monoclonal antibody (Invitrogen, USA) and the avidin-biotin peroxidase complex method with a Vectastain ABC kit (Vector Laboratories, USA). In brief, tissue sections were deparaffinized with xylene and rehydrated through grade alcohols, followed by antigen retrieval in 0.01 M sodium citrate buffer (pH 6.0) and quenching of endogenous peroxidase activity with 3% H2O2. Then, sections were blocked with normal horse serum solution, followed by incubation with anti-SRSF3 primary antibody, biotinalyed anti-mouse IgG, and avidin-biotin peroxidase complex (ABC). Specific staining of SRSF3 was developed by DAB (Vector Laboratories Inc., USA).
SRSF3 specific staining was quantified by using imageJ software [20]. A mean staining value from three representative regions of each sample was calculated. SRSF3 specific staining score was divided to 4 levels (0-3) based on staining value (0: staining value <10, 1: staining value10-15, 2: staining value 15-20, 3: staining value >20).
Oncomine cancer microarray database analyses
Oncomine is an online cancer microarray database [21]. There were nine analyses of oral squamous cell carcinoma vs normal control samples, and one salivary gland cancer vs normal control until Oct 30, 2015. A meta-analysis was used to compare the expression levels of SRSF3 between cancer and normal samples.
DMBA treatment in normal epithelial cells
Normal gingival epithelial cells (N1 and N2) were collected from gingival tissues and grown in keratinocyte growth medium (KGM, Lonza, Switzerland). N2 cells were counted. The same number cells were treated with 5 µg/ml 7, 12-Dimethylbenz[a]anthracene (DMBA) (Sigma-Aldrich, USA) or DMSO (Dimethyl sulfoxide) for two days, and then total proteins were collected with 2× sodium dodecyl sulfate sample buffer.
Western Blot Analysis
Protein samples were separated in 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The blot was incubated with the following antibodies: mouse monoclonal antibodies against SRSF3 (1:100 dilution, Invitrogen, USA), beta-actin (1:1000 dilution, Sigma-Aldrich, USA), and GAPDH (1:1000, Abmart, China).
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was treated with DNase I (Invitrogen), and then reverse-transcribed using random hexamers at 37°C with the M-MLV reverse transcriptase (Promega, Madison, USA). Real-time quantitative RT-PCR was performed using SYBR Green (GeneCopoeia, USA) in a Real-time PCR machine (QuantStudio 6 Flex System, Applied Biosystems, Foster City, CA). Primers for qRT-PCR are synthesized based on the publication [22], except N-cadherin forward primer (5' CCACCTACAAAGGCAGAAGAGA 3'). GAPDH was used as a reference and amplified with following primer pairs: 5'GAAGGTGAAGGTCGGAGTC 3' and 5' GAAGATGGTGATGGGATTTC 3'. The relative levels of gene expression were calculated as ΔCt = Ct(gene) - Ct(reference). The 2-ΔΔCt method was used to calculate the fold-change of gene expression.
Statistical Analyses
The scores of SRSF3 were compared between groups using the nonparametric Mann-Whitney U-test or Kruskal-Wallis test in SPSS software. p<0.05 was considered statistically significant. For real-time qRT-PCR, two group statistical comparisons of means were calculated with student's t-test.
Results
The expression of SRSF3 in OSCC and Dysplasia
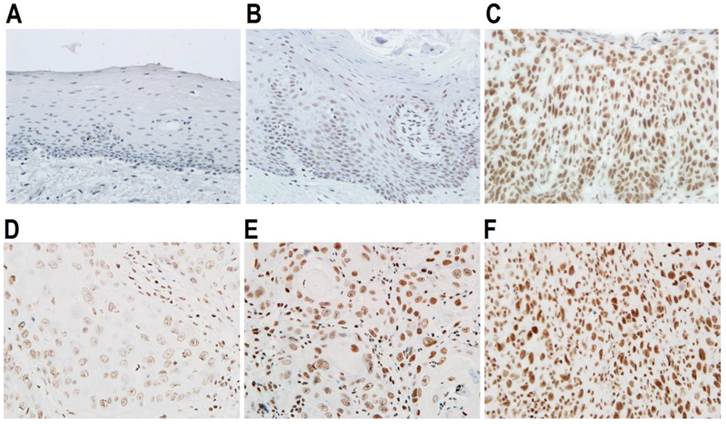
SRSF3 has been shown to be overexpressed in multiple cancers [18]. To understand the potential role of SRSF3 in OSCC, firstly, we compared the expression levels of SRSF3 in normal oral mucosal, dysplasia, and OSCC tissues. We found that SRSF3 is significantly differently expressed in normal, precancerous (dysplasia) and cancerous tissues (p<0.05, Table 1). Cancerous tissues showed significantly higher levels of SRSF3 than normal tissues (p<0.01). Next, we tried to correlated SRSF3 expression levels with the grading of oral epithelial dysplasia. In the early stage of dysplasia (mild stage), there is no significant increased expression of SRSF3. When progressing to moderate or severe dysplasia, lesion tissues showed significantly higher levels of SRSF3 than normal tissues (p<0.01, Table 2, Figure 1A-C). Moreover, we tried to correlate SRSF3 expression levels with the grading of OSCC. We found that SRSF3 is significantly differently expressed in different grades of OSCC (p<0.05, Table 3, Figure 1D-F). OSCCs with higher grade tend to have higher levels of SRSF3.
Expression of SRSF3 in normal, dysplasia, or OSCC tissues.
| Histopathological Classification | Case number (%) | SRSF3 staining | p-value | |||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||
| Normal oral mucosa | 14 (13.7%) | 10 | 3 | 1 | 0 | 0.012 |
| Dysplasia | 11 (10.8%) | 3 | 5 | 2 | 1 | |
| OSCC | 77 (75.5%) | 24 | 25 | 20 | 8 | |
Correlation of SRSF3 expression with the grading of oral epithelia dysplasia.
| Classification | Case number (%) | SRSF3 staining | p-value | ||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||||
| Normal oral mucosa | 14 (13.7%) | 10 | 3 | 1 | 0 | ||
| Dysplasia | Mild | 5 (45.5%) | 3 | 1 | 1 | 0 | 0.53a |
| Moderate to severe | 6 (54.5%) | 0 | 4 | 1 | 1 | 0.006b | |
| Sex (dysplasia) | Male | 6 (54.5%) | 1 | 2 | 2 | 1 | 0.121 |
| Female | 5 (45.5%) | 2 | 3 | 0 | 0 | ||
| Age (displasia) | ≤55 | 4 (36.4%) | 1 | 2 | 1 | 0 | 0.927 |
| >55 | 7 (63.6%) | 2 | 3 | 1 | 1 | ||
a, Normal vs mild dysplasia, b, Normal vs moderate to severe dysplasia.
Expression of SRSF3 in oral epithelial dysplasia and cancers. Expression of SRSF3 was analyzed with immunohistochemistry in normal (A), mild dysplasia (B), severe dysplasia (C), grade I OSCC (D), grade I-II OSCC (E), or grade II-III OSCC tissues (F). (A-F: magnification ×40).
Correlation of SRSF3 expression with clinicopathologic characteristics of OSCC patients.
| Classification | Case number (%) | SRSF3 staining | p-value | ||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||||
| Tumor grade | I | 12 (15.6%) | 8 | 3 | 1 | 0 | 0.016 |
| I-II & II | 49 (63.6%) | 11 | 18 | 16 | 4 | ||
| II-III & III | 16 (20.8%) | 5 | 5 | 2 | 4 | ||
| Lymphatic metastasis | Positive | 38 (49.3%) | 10 | 8 | 15 | 5 | 0.038 |
| Negative | 39 (50.7%) | 16 | 14 | 5 | 4 | ||
| Gender | Male | 58 (75.3%) | 17 | 19 | 13 | 9 | 0.550 |
| Female | 19 (24.7%) | 6 | 7 | 5 | 1 | ||
| Age | ≤55 | 38 (49.4%) | 13 | 14 | 6 | 5 | 0.298 |
| >55 | 39 (50.6%) | 11 | 10 | 13 | 5 | ||
| Tobacco Smoking | Positive | 41 (53.2%) | 11 | 15 | 9 | 6 | 0.59 |
| Negative | 36 (46.8%) | 13 | 9 | 11 | 3 | ||
| Drinking | Positive | 34 (44.2%) | 7 | 12 | 10 | 5 | 0.110 |
| Negative | 43 (55.8%) | 17 | 12 | 10 | 4 | ||
We also used Oncomine microarray database to analyze the expression level of SRSF3 in other studies of oral cancer. A meta-analysis combined nine analyses of oral squamous cell carcinoma vs normal control samples showed significantly higher expression of SRSF3 level in cancer than normal tissues (p<0.001, Figure S1A). A study of salivary gland cancer vs normal control also showed significantly overexpressed SRSF3 in cancer (Figure S1B). These results suggested that SRSF3 is associated with carcinogenesis and progression of oral squamous cell carcinoma.
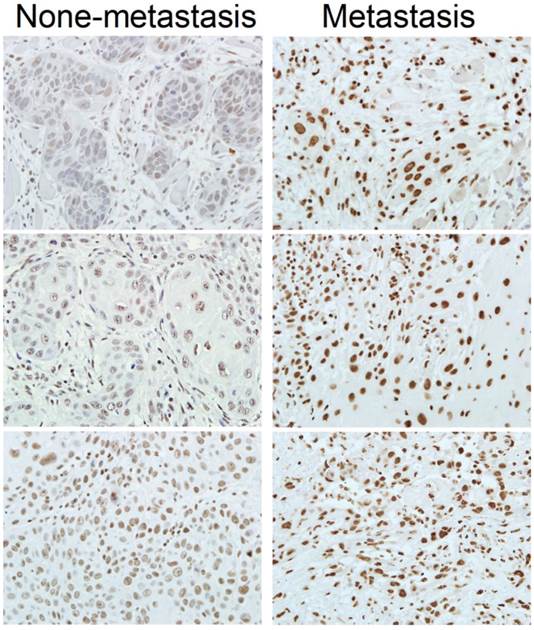
Expression of SRSF3 is associated with lymphatic metastasis of OSCC. Expression of SRSF3 was analyzed with immunohistochemistry in OSCC patients with or without lymphatic metastasis (magnification ×40).
SRSF3 expression is associated with clinicopathologic characteristics of OSCC patients
Metastasis is one of the major factors causing the death of cancer. We found that patients with lymphatic metastasis showed significantly higher SRSF3 expression than those without lymphatic metastasis (p=0.038) (Table 3, Figure 2). This result suggested that SRSF3 might be also involved in metastasis.
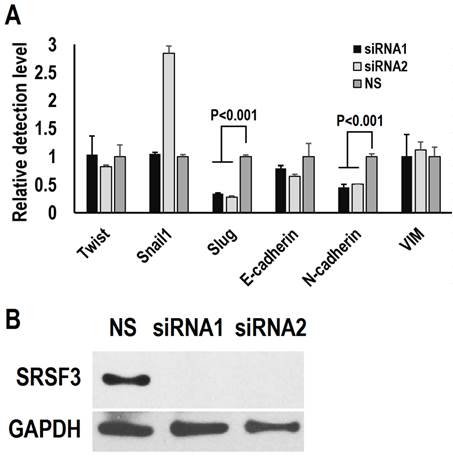
Epithelial-mesenchymal transition (EMT) has been proposed to initiate the metastasis of cancer cells [23, 24]. We investigated the effect of SRSF3 on the expression of EMT related-genes. We knocked down SRSF3 in FaDu cells (a head and neck cancer cell line with metastasis characteristics) with two siRNAs targeting different regions of SRSF3 mRNA (Figure 3B). The expression levels of Slug and N-cadherin significantly decreased after SRSF3 knockdown (Figure 3A). No significantly changes were found in the expression levels of VIM, Snail1, Twist, and E-cadherin. These results indicated that SRSF3 might participate metastasis via regulating the expression of Slug and N-cadherin.
Smoking and drinking are general risk factors of carcinogenesis. We found that there is no significant difference in the expression of SRSF3 level between smoking and non-smoking or between drinking and non-drinking patients (Table 3). SRSF3 may be a relative independent factor during carcinogenesis. In addition, there was no correlation between SRSF3 and age or sex of patients (Table 3).
Carcinogen increased the expression of SRSF3
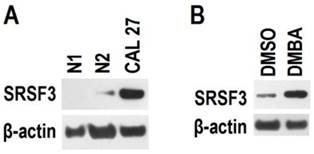
SRSF3 is up-regulated in moderate or severe dysplasia tissues. Many carcinogens are capable to induce dysplasia. Therefore, we analyzed the effect of carcinogen DMBA on the expression of SRSF3. Primary cultured oral epithelial cells had much lower level of SRSF3 than CAL 27 cells (an oral cancer cell line) (Figure 4). When primary cultured oral epithelial cells were exposed to DMBA for two days, SRSF3 expression level significantly increased. This result suggested that carcinogen might be the cause of overexpression of SRSF3.
SRSF3 regulates the expression of epithelial-mesenchymal transition (EMT) related-genes. FaDu cells were treated with siRNA twice in an interval of 48 hours. (A) The expression of E-cadherin (E-cad), Slug, Twist, Snail and Vimentin (VIM) was analyzed by real-time quantitative RT-PCR. GAPDH served as the reference. (B) Western blot showed knockdown efficiency of SRSF3 in FaDu cells. GAPDH served as loading control.
Discussion
Oral squamous cell carcinoma, encompassing more than 90% of oral tumors, counts among the most frequent malignancies of head and neck [1]. Five-year relative survival rate (%) of oral cancer patients, diagnosed between 2003 and 2009, is 65%, which is much lower than those of breast cancer patients and prostate cancer patients [2]. An important reason for poor treatment outcome of OSCC is the poor understanding of its tumorigenesis. OSCC is associated with aberrant alternative splicing of pre-mRNA due to overexpression of splicing factor hnRNP A1 [6]. Here, we focused on another key splicing factor, SRSF3, and expanded our knowledge of how splicing factor is associated with OSCC.
Previous studies showed that SRSF3 is overexpressed in cancers of the cervix and ovary [18, 25]. Recently, in consistent with previous studies, we found that SRSF3 is overexpressed in OSCC cells [19]. Here, we also found that SRSF3 was overexpressed in OSCC tissues based on a meta-analysis of oral cancer researches in Onocmine microarray database. In this study, we further investigated the relationship between SRSF3 expression and clinical characteristics of OSCC patients. Because most of our samples are formalin-fixed, paraffin-embedded tissues, we only performed immunohistochemistry to analyze the expression of SRSF3 in patients instead of qRT-PCR or Western blot. We found that SRSF3 is up-regulated in moderate or severe dysplasia tissues compared with normal oral mucosal epithelial tissues, and higher grade cancers express more SRSF3. Our data clearly showed gradually increased expression of SRSF3 is associated with the development of precancerous lesions and cancer. SRSF3 may be a new potential molecular marker for OSCC.
Carcinogen increased the expression of SRSF3. (A) Expression of SRSF3 in primary cultured oral epithelial cells (N1 and N2) and CAL 27 (an oral cancer cell line) was analyzed with western blot. (B) Primary cultured oral epithelial cells (N2) were treated with DMBA (5 µg/ml) for two days. DMSO treatment was used as control. Western blot was used to analyze the expression of SRSF3. β-actin served as loading control.
Overexpression of SRSF3 in an immortalized mouse embryonic fibroblast has been demonstrated to be able to initiate tumor formation [18]. We found that carcinogen DMBA significantly increased the expression of SRSF3 in normal oral primary epithelial cells. In addition, moderate or severe dysplasia tissues showed significantly higher SRSF3 than normal tissues. These results are correlated with previous study and strongly suggest that SRSF3 may be involved in the initiation of OSCC and be a therapeutic target for the prevention of OSCC.
Metastasis is the major cause of cancer related mortality [26]. Splicing factor SRSF1 has been reported to enhance metastasis by promoting the expression of ΔRon isoform of Ron tyrosine kinase receptor [27]. Here, we proposed that SRSF3 is associated with metastasis based on two evidences: first, patients with lymphatic metastasis showed significantly higher SRSF3 expression than those without lymphatic metastasis; second, knockdown of SRSF3 significantly reduced the expression of two EMT-related genes, Slug and N-cadherin. Slug represses the expression of E-cadherin [28]. However, we did not observe the increase of E-cadherin upon SRSF3 knockdown. This may be because that the expression of E-cadherin is also repressed by Snail [29]. In the present study, we found that one anti-SRSF3 siRNA has no effect on Snail, whereas another anti-SRSF3 siRNA increased the expression of Snail. Therefore, SRSF3 only regulates the expression of Slug.
N-cadherin has been shown to be associated with anti-apoptotic signaling pathways [30]. Its transcription is activated by Twist [31]. Our results showed decrease of N-cadherin and no change in Twist upon SRSF3 knockdown. This may suggest that SRSF3 regulates the expression of N-cadherin via Twist-independent pathway.
Alternative splicing has been increasingly associated with cancers [32]. SR splicing factor family (such as SRSF3 [18] and SRSF1 [33]) and SR-related proteins (such as Tra2β) [34] have been concluded to be oncogene or closely associated with oncogenesis. However, the relationship between these splicing factors and carcinogenesis and caner progression remains largely unclear. In the present study, we provided more evidences about how SRSF3 is correlated with progression of OSCC. Further studies may focus on the therapeutic strategies targeting SRSF3 in cancers.
Conclusions
In conclusion, we found that SRSF3 is overexpressed in OSCC tissues and associated with the initiation and development of precancerous lesions and cancer. SRSF3 may be a biomarker and therapeutic target of OSCC.
Supplementary Material
Figure S1.
Abbreviations
OSCC: Oral squamous cell carcinoma; SR proteins: serine/arginine-rich proteins; RRM: RNA recognition motif; DMEM: Dulbecco's modified Eagle medium; ABC: avidin-biotin peroxidase complex; DMBA: 7, 12-Dimethylbenz[a]anthracene; DMSO: Dimethyl sulfoxide; qRT-PCR: Real-time quantitative RT-PCR; EMT: Epithelial-mesenchymal transition.
Acknowledgements
This work was supported by Grant 81271143 and 81470741 from the National Science Foundation of China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin. 2002;52:195-215
2. Siegel R, Ma J, Zou Z. et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29
3. Shah TM, Patel AK, Bhatt VD. et al. The landscape of alternative splicing in buccal mucosa squamous cell carcinoma. Oral Oncol. 2013;49:604-10
4. Palve V, Mallick S, Ghaisas G. et al. Overexpression of Mcl-1L splice variant is associated with poor prognosis and chemoresistance in oral cancers. PLoS One. 2014;9:e111927
5. Ishii H, Saitoh M, Sakamoto K. et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem. 2014;289:27386-99
6. Yu C, Guo J, Liu Y. et al. Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J Cell Physiol. 2015;230:2252-61
7. Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010;24:1073-4
8. Zahler AM, Lane WS, Stolk JA. et al. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837-47
9. Cavaloc Y, Bourgeois CF, Kister L. et al. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468-83
10. Majerciak V, Lu M, Li X. et al. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA. 2014;20:1747-58
11. He X, Zhang P. Serine/arginine-rich splicing factor 3 (SRSF3) regulates homologous recombination-mediated DNA repair. Mol Cancer. 2015;14:158
12. Huang Y, Gattoni R, Stevenin J. et al. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837-43
13. Escudero-Paunetto L, Li L, Hernandez FP. et al. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology. 2010;401:155-64
14. Lou H, Neugebauer KM, Gagel RF. et al. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977-85
15. Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459-67
16. Kim J, Park RY, Chen JK. et al. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5'-UTR region. Cell Death Differ. 2014;21:481-90
17. Corbo C, Orru S, Salvatore F. SRp20: an overview of its role in human diseases. Biochem Biophys Res Commun. 2013;436:1-5
18. Jia R, Li C, McCoy JP. et al. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6:806-26
19. Guo J, Jia J, Jia R. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci Rep. 2015;5:14548
20. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671-5
21. Rhodes DR, Yu J, Shanker K. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1-6
22. Scheel C, Eaton EN, Li SH. et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926-40
23. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442-54
24. Thiery JP, Acloque H, Huang RY. et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
25. He X, Arslan AD, Pool MD. et al. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene. 2011;30:356-65
26. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70
27. Ghigna C, Giordano S, Shen H. et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881-90
28. Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613-8
29. Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155-66
30. Tran NL, Adams DG, Vaillancourt RR. et al. Signal transduction from N-cadherin increases Bcl-2. Regulation of the phosphatidylinositol 3-kinase/Akt pathway by homophilic adhesion and actin cytoskeletal organization. J Biol Chem. 2002;277:32905-14
31. Alexander NR, Tran NL, Rekapally H. et al. N-cadherin gene expression in prostate carcinoma is modulated by integrin-dependent nuclear translocation of Twist1. Cancer Res. 2006;66:3365-9
32. Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228-37
33. Karni R, de Stanchina E, Lowe SW. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185-93
34. Best A, Dagliesh C, Ehrmann I. et al. Expression of Tra2 beta in Cancer Cells as a Potential Contributory Factor to Neoplasia and Metastasis. Int J Cell Biol. 2013;2013:843781
Author contact
![]() Corresponding authors: Rong Jia, 237 Luoyu Road, Wuhan City, 430079, PR China. Tel: +862787686268. E-mail address: jiarongedu.cn Or Jihua Guo, 237 Luoyu Road, Wuhan City, 430079, PR China. Tel: +862787686208. E-mail address: jihuaguoedu.cn
Corresponding authors: Rong Jia, 237 Luoyu Road, Wuhan City, 430079, PR China. Tel: +862787686268. E-mail address: jiarongedu.cn Or Jihua Guo, 237 Luoyu Road, Wuhan City, 430079, PR China. Tel: +862787686208. E-mail address: jihuaguoedu.cn

 Global reach, higher impact
Global reach, higher impact