Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(6):424-431. doi:10.7150/ijms.14475 This issue Cite
Research Paper
Detection of Human Papillomavirus 16-Specific IgG and IgM Antibodies in Patient Sera: A Potential Indicator of Oral Squamous Cell Carcinoma Risk Factor
1. Department of Oral Biology & Biomedical Sciences, Faculty of Dentistry, University of Malaya, 50603, Kuala Lumpur, Malaysia
2. Institute of Nano Electronic Engineering (INEE), Universiti Malaysia Perlis (UniMAP), 01000, Kangar, Perlis, Malaysia
3. School of Bioprocess Engineering, Universiti Malaysia Perlis (UniMAP), 02600 Arau, Perlis, Malaysia.
4. Faculty of Accountancy and Management, University Tungku Abdul Rahman, 43000, Kajang, Selangor, Malaysia
5. Advanced Medical & Dental Institute (AMDI), Universiti Sains Malaysia, 13200, Kepala Batas, Pulau Pinang, Malaysia
6. Biomaterial Research Laboratory, Dental Research Management Centre, Faculty of Dentistry, University of Malaya, 50603, Kuala Lumpur, Malaysia
7. Department of Oro-Maxillofacial Surgical & Medical Sciences, Faculty of Dentistry Building, University of Malaya, 50603, Kuala Lumpur, Malaysia
8. Oral Cancer Research and Coordinating Centre, Faculty of Dentistry, University of Malaya, 50603, Kuala Lumpur, Malaysia
Received 2015-11-18; Accepted 2016-4-21; Published 2016-5-12
Abstract
The association between human papillomavirus type 16 (HPV16) and oral cancer has been widely reported. However, detecting anti-HPV antibodies in patient sera to determine risk for oral squamous cell carcinoma (OSCC) has not been well studied. In the present investigation, a total of 206 OSCC serum samples from the Malaysian Oral Cancer Database & Tissue Bank System, with 134 control serum samples, were analyzed by enzyme-linked immunosorbant assay (ELISA) to detect HPV16-specific IgG and IgM antibodies. In addition, nested PCR analysis using comprehensive consensus primers (PGMY09/11 and GP5+/6+) was used to confirm the presence of HPV. Furthermore, we have evaluated the association of various additional causal factors (e.g., smoking, alcohol consumption, and betel quid chewing) in HPV-infected OSCC patients. Statistical analysis of the Malaysian population indicated that OSCC was more prevalent in female Indian patients that practices betel quid chewing. ELISA revealed that HPV16 IgG, which demonstrates past exposure, could be detected in 197 (95.6%) OSCC patients and HPV16-specific IgM was found in a total of 42 (20.4%) OSCC patients, indicating current exposure. Taken together, our study suggest that HPV infection may play a significant role in OSCC (OR: 13.6; 95% CI: 3.89-47.51) and HPV16-specific IgG and IgM antibodies could represent a significant indicator of risk factors in OSCC patients.
Keywords: Human Papillomavirus 16, Oral Squamous Cell Carcinoma, Enzyme-Linked Immunosorbant Assay, Nested PCR, Oral cancer
Introduction
Oral cancer is the sixth most prevalent malignancy worldwide, causing severe illness that leads to a significant death rate. The incidence of oral cancer varies from country to country or region to region, depending on personal habits, awareness and prevention strategies. In fact, when considering Peninsular Malaysia from 2003 to 2005, the National Cancer Registry (NCR) reported oral cancer as the 22nd and 15th most common cancer type among males and females, respectively [1]. Even with modern medical treatments, it has been estimated that the overall mortality rate for oral cancer remains high (approximately 50%) [2]. This cancer has been associated with various factors, with incidences rates linked to the use of tobacco, betel quid chewing, and alcohol consumption [3]. However, it has also been reported that oral cancer may develop in the absence of exposure to these risk factors, but with other related factors including diet, ionizing radiation, genetic predisposition and viruses of the oral cavity (e.g., human papillomavirus [HPV]) [4, 5]. A fundamental pitfall with regard to the survival rate of OSCC patients is that, most cases are diagnosed in the advanced stage. In this regard, it is fundamental that assessments be developed for the easy identification of risk factors. For instance, in a recent study of proteomic analyses on sera from OSCC patients in Malaysia, researchers identified various biomarkers that can be used in the diagnosis and prognosis of OSCC [6]. In addition, since microbial detection probes are easily employed, cancer-associated microbs (e.g., HPV) could be used in the assessments/diagnosis of malignancies.
One of the reported risk factor of oral cancer is human papillomavirus (HPV). HPV can be classified into either low-risk or high risk sub-types based on their presence in malignant lesions. There are nearly 200 different HPV sub-types that have been reported. However, 20 of these HPV sub-types are known to be linked to cancer risk, and high risk types (e.g., 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68) have been demonstrated to be involved in epithelial carcinogenesis [7, 8]. In fact, according to the International Agency for Research on Cancer, high-risk HPV (HR-HPV) types 16 is classified as the biological agent which is carcinogenic to humans, and is responsible for the development of various cancers such as uterine cervix cancer [9, 10]. High-risk HPVs have also been proposed as etiological factors for various head and neck squamous cell carcinomas (HNSCC), tonsillar cancers, and OSCC [9], and these high-risk HPVs are reported to be most detected (59%) in the oral cavity cancer [11]. Apart from these, studies have shown, women with previous cervical HPV infection have higher oral HPV prevalence compared to the others [12]. In this respect, the majority of studies have focused on HPV16, as it is more significantly associated with oral cavity cancer [5, 13] . Due to the high incidence rates of HPV infections in oral cancers patients [14], it is important to investigate the association of HPV with oral cancer. As previously mentioned, there exist various probes and antibodies that have been widely used for decades in the detection of cancer. In this study, the presence of antibodies against HPV is evaluated as a risk factor for OSCC.
In general, during human immune response to HPV, B cells detect the viral antigens and exhibit them to T helper type 2 cells, which in turn promote the production of high-affinity antibodies (immunoglobulin G, A and M [IgG, IgA and IgM]) against HPV antigens by B cells. Indeed, it has already been demonstrated that anti-HPV IgG could be measured by enzyme-linked immunosorbant assay (ELISA), representing a reliable marker for past HPV exposure [15]. Also, a study by Harro et al. [16] on HPV16 vaccination indicated that most vaccine recipients became seropositive with HPV16 IgM, which was typically detected 1 month after initial immunization. Thus, the presence of HPV16 IgM antibodies directly shows acute or current exposure of HPV. On the other hand, the presence of HPV16 IgG antibodies represents past exposure to HPV. Studies have shown that the median duration for HPV16 IgG was 3 years [17]. As indicated by Petter et al. [18] serological assays for HPV could help to identify those patients at risk of HPV-related cancers. In addition to antibody-based detection strategies, DNA sequencing or PCR method are also widely used in the detection of viral DNA of HPV in tissue samples [19]. Therefore, antibody- and DNA-based assays can complement each other for the reliable identification of HPV-infected patients.
In the present study, we have analyzed serum samples collected from OSCC from Malaysian Oral Cancer Database & Tissue Bank System (MOCDTBS) [20] at the Oral Cancer Research & Coordinating Centre (OCRCC), University of Malaya (UM). In particular, we have used indirect ELISA to measure HPV16-specific IgG and IgM antibodies in order to determine whether the presence of these HPV IgG and IgM antibodies could be used as an OSCC risk indicator. Moreover, nested PCR (nPCR) was used to validate the presence of specific HPV DNA sequences in OSCC samples using two different consensus primers from distinct regions of the HPV virus. To support our experimental evidences, data related to demographics and personal habits (tobacco smoking, alcohol consumption and betel quid chewing) that might be associated with HPV-infected OSCC patients were also collected and considered as causal factors.
Materials and Methods
Study Population and Sample Collection
A total of 206 OSCC sera samples collected by the MOCDTBS at OCRCC (UM) were used in our serological analyses. Also, genomic DNA samples from fresh frozen OSCC tissue corresponding to 84 of the 206 samples above were used for HPV detection and typing by nPCR. Sera from 134 healthy individuals were also analyzed as the control group. All samples were collected with informed consent before recruitment, and approval for the study was obtained from the Medical Ethics Committee for the Faculty of Dentistry, University of Malaya (ref no: DF DR1307/0077(U)). Due to the un-available record on the patients HPV vaccine immunization, all patients were considered to be HPV vaccine free since patients were diagnosed and recruited before the introduction of the HPV vaccine in Malaysia.
HPV Serological Analyses
IgG and IgM against HPV16 antigens were detected in patient sera using HPV16 antibody ELISA kits according to the manufacturer's protocols (Cusabio, Wuhan, China). These kits included microtiter plates that were pre-coated with HPV-specific antigens. Uniformly diluted serum samples were added to the microtiter wells and incubated for 30 min at 37°C. The wells were subsequently washed, and horseradish peroxidase (HRP)-conjugated anti-human IgG/IgM was added and incubated again for 30 min at 37°C. After thorough washing, 3,3',5,5' tetramethylbenzidine (TMB) substrate solution was added to each well, and the enzyme-substrate reaction was terminated by the addition of sulphuric acid solution. The color changes were then measured spectrophotometrically at 450 nm using a microplate reader (Tecan Infinite m200 Pro, Tecan Group Ltd., Mannedorf). The valence of HPV16 antibody (IgG or IgM) in the samples was detected through optical density (OD) based on the manufacturer's protocol. In this regard, the cutoff level for HPV16 seropositivity was determined according to the instructions provided by the manufacturer and the positive and negative control was provided with the kit.
HPV Detection Using Nested PCR
In order to perform a thorough and precise PCR analysis, OSCC DNA sample preparation, PCR reagent preparation/setup, and PCR product analysis were performed in three separate rooms using dedicated instruments. A total of 84 genomic DNA samples were obtained from the MOCDTBS. To exclude false negative results, the quality of the extracted DNA specimens was determined by analyzing 5 μL of each DNA sample in a PCR assay targeting a 268-bp region of the β-globin-specific gene using the following primers: PC04 and GH20 (Table 1) [21]. The PCR products were then visualized by 2% agarose gel electrophoresis. DNA amplification for HPV detection was then performed via nPCR on all of the β-globin-positive DNA samples.
To screen for the presence of HPV in the OSCC samples, we first used a primary PCR procedure with the L1 consensus PCR primer pools PGMY09/11 (primary PCR) and primer HMB01 (Table 1) targeting the 450-bp region. This procedure was based on previously described protocols [22-24], which were modified and optimized. Briefly, 1 μL (30 - 50 ng) of DNA sample was amplified with an equimolar mixture of the primers (i.e., PGMY09 and PGMY11; final concentration of 10 pmol for each). The DNA sample and primer mixture was complemented with an optimal concentration of PCR Buffer containing 2 mM MgCl2, PCR grade deoxyribonucleoside triphosphates / dNTP mix (10 mM of each nucleotide), 2U of FastStart Taq DNA Polymerase (Roche, Germany), and nuclease free water. Amplification was performed with an Applied Biosystems® Veriti® 96-Well Thermal Cycler (USA). The PCR cycling conditions for the consensus primers (PGMY09/11) were as follows: 95°C for 5 min (initial denaturing), followed by 40 cycles of 95°C for 1 min (denaturing), 60°C for 1 min (annealing), and 72°C for 1 min (elongation). This was followed by a final extension period of 10 min at 72°C and storage at 4°C.
Primers used to detect HPV in clinical samples.
| Primer Set | Primer Name | 5'-3' sequence |
|---|---|---|
| β-globin | GH2O | GAA GAG CCA AGG ACA GGT AC |
| PCO4 | CAA CTT CAT CCA CGT TCA CC | |
| PGMY09/11 | PGMY11-A | GCA CAG GGA CAT AAC AAT GG |
| PGMY11-B | GCG CAG GGC CAC AAT AAT GG | |
| PGMY11-C | GCA CAG GGA CAT AAT AAT GG | |
| PGMY11-D | GCC CAG GGC CAC AAC AAT GG | |
| PGMY11-E | GCT CAG GGT TTA AAC AAT GG | |
| PGMY09-F | CGT CCC AAA GGA AAC TGA TC | |
| PGMY09-G | CGA CCT AAA GGA AAC TGA TC | |
| PGMY09-H | CGT CCA AAA GGA AAC TGA TC | |
| PGMY09-I | G CCA AGG GGA AAC TGA TC | |
| PGMY09-J | CGT CCC AAA GGA TAC TGA TC | |
| PGMY09-K | CGT CCA AGG GGA TAC TGA TC | |
| PGMY09-L | CGA CCT AAA GGG AAT TGA TC | |
| PGMY09-M | CGA CCT AGT GGA AAT TGA TC | |
| PGMY09-N | CGA CCA AGG GGA TAT TGA TC | |
| PGMY09-P | G CCC AAC GGA AAC TGA TC | |
| PGMY09-Q | CGA CCC AAG GGA AAC TGG TC | |
| PGMY09-R | CGT CCT AAA GGA AAC TGG TC | |
| HMB01 | GCG ACC CAA TGC AAA TTG GT | |
| GP5+/GP6+ | GP5+ | TTT GTT ACT GTG GTA GAT ACT AC |
| GP6+ | GAA AAA TAA ACT GTA AAT CAT ATT C |
a PGMY09-I and PGMY09-P are 18 bp in length. The first two 59 bases were deleted to reduce the significant internal secondary structure of the oligonucleotide
b HMB01 is shifted 39 from the downstream primer region of the other HPV genotypes to avoid secondary structure formation and internal priming.
The secondary PCR involved amplification of 1 μL of the primary PCR product using the general consensus primer GP5+/6+ (Table 1), which targets a 150-bp fragment based on modification of a previously described protocol [25-27]. The reagents, buffer, and instrument used in this PCR amplification were identical to that of the primary PCR protocol. The PCR cycling conditions for the consensus primers were as follows: 94°C for 120 s (initial denaturing); followed by 40 cycles of 94°C for 45 s (denaturing); 48°C for 4 s, 38°C for 30 s, 42°C for 5 s, 66°C for 5 s (annealing); and 71°C for 90 s (elongation). This was followed by a final extension period of 10 min at 72°C and storage at 4°C. All PCR analyses were performed along with a positive control (DNA from HPV-positive HeLa cells) [28], and a non-template control was used to evaluate contamination and accuracy (negative control).
The PCR products for β-globin as well as the primary and secondary PCR reactions were electrophoresed for 35 min at 110V using 2% low melting point agarose gels (Vivantis Inc., USA) in 1X Tris-borate-EDTA (TBE) buffer. The gels were then stained with ethidium bromide, visualized, and photographed (MultiDoc-It Imaging System, UVP, Upland, CA). As a measure for quality control, HPV detection was randomly repeated on some samples and compared to the initial results.
Statistical Analysis
The association between risk factors and HPV were analyzed using SPSS 12.0.1. A p-value < 0.05 was considered to be statistically significant. Logistic regression was conducted to assess whether the predictors (i.e., HPV16 antibodies, gender, race, and age) were significantly associated with OSCC. The assumptions related to independent errors, multicollinearity, normality, nomoscedasticity, and outliers were checked and met.
Results
Patients Characteristics
This investigation included a total of 208 OSCC patients (cases) and 134 non-OSCC patients (control) with a mean age of 48.9±17.4. Notably, two patient samples were excluded from the study due to inadequate clinical and demographic information. Based on the patients socio-demographic profiles, female (67.0%) Indians (49.5%) shows the highest number of patients diagnosed with OSCC. Tobacco smoking, alcohol drinking and betel quid chewing are the most common risk habits in OSCC population. However, betel quid chewing (43.7%) remains the highest possible risk factor in the development of OSCC among the Malaysian population (Table 2).
Serological Analysis
HPV seropositivity of OCSS patients and control was evaluated using an HPV16-specific ELISA assay to detect both Immunoglobulin G (IgG) and Immunoglobulin M (IgM). Based on these ELISA analysis, 197/206 OSCC patients and 89/134 control were positive for HPV IgG, whereas only 42/206 of OSCC patients were seen positive for HPV 16 IgM (Table 3).
Social habits (etiologic risk factors of OSCC) of patients (n=206) with OSCC
| Social Habits | No. of Patients | % |
|---|---|---|
| Tobacco Smoking | ||
| Smoker | 48 | 23.3 |
| Non-Smoker | 136 | 66.0 |
| Not- Available | 22 | 10.7 |
| Alcohol Drinking | ||
| Alcoholic | 50 | 24.3 |
| Non-Alcoholic | 134 | 65.0 |
| Not- Available | 22 | 10.3 |
| Betel Quid Chewing | ||
| Chewing | 90 | 43.7 |
| Not-Chewing | 94 | 45.6 |
| Not- Available | 22 | 10.7 |
Percentage of distribution for HPV16 IgG and HPV16 IgM among the OSCC patients and the control group
| OSCC Samples (n=206) | Control Samples (n=134) | |||
|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | |
| HPV16 IgG | ||||
| Positive | 197 | 95.6 | 89 | 66.4 |
| Negative | 9 | 4.4 | 45 | 33.6 |
| HPV16 IgM | ||||
| Positive | 42 | 20.4 | 0 | 0 |
| Negative | 164 | 79.6 | 134 | 100 |
Incidence Rate and Prediction of OSCC
To assess whether certain patients characteristic or variables could significantly predicts the likelihood of OSCC, a logistic regression analysis was employed. Based on these analyses, four independent variables (i.e. HPV 16 IgG, gender, race and age) were identified to make a unique statistically significant contribution in predicting the likelihood of OCSS (Table 4). The logistic regression analysis identifies HPV 16 IgG as the strongest predictor, recording an odd ratio of 13.6, followed by female (gender) with the significant ratio of 4.01. In addition Indians (race) showed significant prediction to OSCC when compared with the other race in Malaysia. And lastly, the model predicts that the chances of OSCC were 1.15 times higher with every additional year of age.
HPV Detection Using Nested PCR
A total of 84 genomic DNA were used to detect the presence of HPV in the OSCC samples through nested PCR. The DNA samples used in this study were from the similar patients that were used in the serological assay. The quality of all the genomic samples was screened through PCR assay targeting β-globin-specific gene with the primers PC04 and GH20. All samples gave positive amplification to β-globin-specific gene and were further analyzed through nested PCR (nPCR).
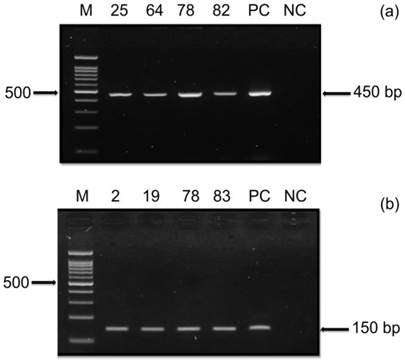
Nested PCR using two general consensus PCR primers (PGMY09/11 and GP5+/6+) was employed to validate the presence of HPV in the OSCC samples. The resulting PCR product was then further examined using agarose gel electrophoresis to determine the presence of near identical bands size to the expected band. Based on the gel electrophoresis, 8/84 DNA samples amplified with PGMY09/11 and 18/84 DNA sample tested with GP5+/6+ were found positive (Table 5).
Logistic Regression analyses in predicting the risk factors in Oral Squamous Cell Carcinoma
| B | SE | Odd Ratio | P-value | 95% C.I for Lower | 95% C.I for Upper | |
|---|---|---|---|---|---|---|
| HPV16 IgG | 2.61 | 0.64 | 13.59** | 0.00 | 3.89 | 47.51 |
| Gender | 1.39 | 0.47 | 4.01** | 0.00 | 1.59 | 10.07 |
| Indian | 0.00 | |||||
| Race (Indian vs Malay) | -2.26 | 072 | 0.10** | 0.00 | 0.03 | 0.43 |
| Race (Indian vs Chinese) | -1.71 | 0.81 | 0.18* | 0.03 | 0.04 | 0.88 |
| Race (Indian vs Others) | 1.11 | 1.71 | 3.04 | 0.52 | 0.11 | 86.71 |
| Age | 0.14 | 0.02 | 1.15** | 0.00 | 1.10 | 1.19 |
| Constant | -6.55 | 1.69 | 0.00 | 0.00 |
Note: R2 = 0.561 (Cox and Snell), 0.774 (Nagalkerke). Model χ2 (7) = 254.3, p<0.001. *p<0.05, **p<0.01.
Frequency percentage of HPV detection using PGMY09/11 and GP5+/6+ nested PCR
| PGMY09/11 | PGMY09/11 / nGP5+/6+ | ||||
|---|---|---|---|---|---|
| No. of patients | % | No. of patients | % | ||
| Positive | 8 | 9.5 | 18 | 21.4 | |
| Negative | 76 | 90.5 | 66 | 78.6 | |
| Total | 84 | 100 | 84 | 100 | |
Discussion
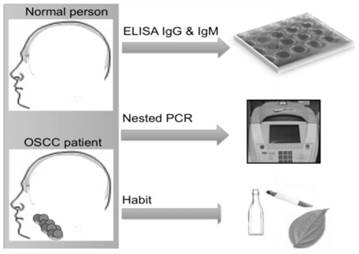
The presence of HPV antibodies indicates infection history, which can occur at various sites in the body, including the genitals and oral cavity. Studies have shown that HPV infection can develop into a malignant state within a certain number of years. In cervical cancer, CIN1 (low-grade lesions) which is a sign of an active HPV-infection, can develop within weeks to few months from HPV exposure and the development of CIN3 could take up to 9.4 years. Thus, it has been reported that cancer occurs after transformation of latently infected cells, which may even take decades to develop [22]. In a recent study by Kreimer and his colleagues [29], the researchers suggested that HPV16 seropositivity was present in a patient's serum for more than a decade before oropharynx cancer was first diagnosed. Therefore, identifying HPV infection and understanding the role of HPV IgG and IgM antibodies might assist in the early detection of HPV-derived cancers. For this reason, the present study focuses on the serological screening of OSCC patient samples, which was further validated by nPCR and studied in the context of social demographic profiles in order to evaluate potential causal factors (Figure 1).
Study design. Sample collected from both normal and OSCC patient. Analyzed for the presence HPV-IgG and HPV-IgM. Confirmation of sequence by nested PCR. Personal habits are analyzed.
HPV Serological Analyses
The ELISA test was developed in 1971 to evaluate the interactions between antibodies and antigens, and due to its simplicity and reliability, it has been widely use in the field of oncology [30-32]. Here, HPV seropositivity of OCSS patients was evaluated using an HPV16-specific ELISA assay. Notably, this HPV genotype is the most commonly associated with head and neck cancer. We used the ELISA assay to detect the presence of both IgG and IgM against HPV (i.e., past and current exposure to HPV, respectively). Based on these analyses, the number of patients with HPV16 IgG was observed to be significantly higher among OSCC patients (95.6%; n=197) and the control group (66.4%; n=89) compared to the number of IgM-positive patients (Table 3). This finding suggest that past exposure to HPV16 infection might contributed to the onset of OSCC, further supporting the hypothesis by Petter et al. [18]. Since HPV IgG serological response is an indicator of past HPV infection, the presence of HPV IgG may better represent the relationship of HPV and OSCC [33-36]. Furthermore, among the 206 OSCC patients, 20.4% (n=42) displayed acute HPV16 infection, which was detected by the presence of HPV-specific IgM in the serum that was 5-fold less than patients with HPV IgG seropositivity. Taken together, this indicates that the role of past infection as a risk factor is predominantly higher in the Malaysian population
Regression Analysis and Prediction
Based on the above data, a logistic regression analysis was employed to assess whether certain patient characteristics (e.g., HPV16 IgG, gender, race, and age) could significantly predict the likelihood of OSCC (Table 4). When all four variables were considered together, they significantly predicted the model (χ2 = 254.3, df = 7; N = 340; p <0.001), indicating that the model was capable of distinguishing the likelihood of OSCC. Indeed, the model as a whole explained between 56.1% (Cox and Snell R-square) and 77.4% (Nagelkerke R-square) of the variance and correctly classified 90.6% of the cases. As shown in Table 4, four independent variables were found to make a uniquely significant contribution to the model, which includes HPV16 IgG, gender, race, and age. However, the strongest predictor in the model was HPV16 IgG, recording an odds ratio of 13.6. This indicates that, based on our data the patients who are positive for HPV16 IgG is 13.6 times more likely to have OSCC when controlling for all other factors in the model. Moreover, our results further emphasized the relationship between HPV seropositivity and OSCC, confirming previous reports [36, 37]. As for gender, females were about four times more likely to have OSCC compared to males (OR=4.01) a findings consistent with the previous study [38]. However, studies have shown that the incidence rates of oral cancer are low among women in several countries because of the differences in lifestyle behaviors between genders [39]. Race also showed significant predictive value for OSCC, with Indians used as the reference due to their high prevalence of OSCC. In this respect, logistic regression predicted that Malays were approximately 1/10 as likely to have OSCC and Chinese were about 1/5 as likely to have OSCC when compared to the Indian population. Suggesting that Malays and Chinese are less likely to have OSCC compared to Indians. In addition, our analysis indicated that the chance of having OSCC was 1.15 times higher with every additional year of age (OR=1.15). Although, oral cancer has been commonly reported in middle-aged and older age group, the occurrence in younger adults has been documented in the recent years [40]. Furthermore, studies have shown that HPV-related OSCC were diagnosed at younger age and the prevalence becomes higher as the age increases [41, 42]. Taken together, the detection of HPV IgG antibodies in the majority of OSCC patient samples (95.6%) suggested that the presence of IgG antibody (OR: 13.59; 95% CI: 3.89-47.51) could be a valuable predictor of OSCC risk. Thus, HPV-specific IgG antibody could function as a surrogate marker for oral carcinogenesis [33] potentially contributing to prevention strategies and/or early diagnosis of OSCC.
Selective Amplification of HPV Sequence by Nested PCR
Current technologies (e.g., in situ hybridization, PCR, and DNA sequencing) are widely used to detect viral DNA/RNA transcripts [19, 43] and to diagnose HPV infection. Nested PCR represents a modified method among the conventional PCR techniques, which are employed to obtain specific amplifications. Thus, to confirm the presence of HPV in our samples, we also performed nPCR on a total of 84 genomic OSCC DNA specimens. In this regard, the genomic DNA samples used in the nPCR were from the same patients analyzed in the serological assay. Notably, the genomic samples were screened in a PCR assay targeting a 268-bp region of the β-globin-specific gene using PC04 and GH20 primers in order to exclude false negative results and to verify the quality of the extracted DNA. All of the samples gave positive results in this β-globin-specific assay and were further analyzed by nPCR.
To further validate the presence of HPV in the OSCC samples used in the serological assay, nPCR using two general consensus PCR primers was performed. The nPCR was first conducted using highly sensitive HPV PGMY09/11 primers (primary PCR), which is the most recent version of MY09/MY11 [22]. This was followed by amplification with GP5+/6+ primers (secondary PCR) using 1 μL of the PCR product from the primary reaction. Agarose gel electrophoresis was then carried out to visualize the corresponding PCR products (Figure 2). Based on the HPV nPCR analysis, 8/84 (9.5%) OSCC samples were found to be positive using the PGMY09/11 primers (primary PCR), whereas 18/84 (21.4%) samples were observed to be positive with the PGMY09/11 and GP5+/6+ primer set (secondary PCR) (Table 5). Thus, our overall HPV detection rate was 28.6% in OSCC using both PGMY09/11 and nested GP5+/6+ primers, which is in line with the rate (22.8%) reported in a previous study by Yang et al. [44]. Nested PCR using the combined PGMY09/11 and GP5+/6+ primer set was able to detect a higher frequency of HPV-positive samples than the primary PCR. This is because nPCR displays a higher sensitivity for detecting HPV compared to one-step PCR [45]. Nevertheless, the chances of obtaining a false negative result with this method should not be ruled out, which could be due to the fact that oral samples may produce weaker PCR products when compared to HPV-positive cervical specimens [46]. However, based on our findings, the detection of HPV antibodies was not in strict concordance with HPV DNA results. This is because antibody responses (mainly IgG) give a general indication of past exposure, whereas HPV DNA detection reflects on the current presences of HPV in the cancer tissue.
HPV Nested PCR. The presence of HPV in the OSCC samples were tested using (a) HPV primer PGMY09/11 and (b) nested HPV primer GP5+/6+. A representative gel is shown from 84 samples tested with the two consensus primers displaying positive PCR amplification for PGMY09/11 (450bp) and nested GP5+/6+ (150bp). Positive Control (PC) using DNA from HeLa cells and negative control (NC) were included in the experiments. Marker and sample ID are indicated.
Social Habits of OSCC Patients
In order to understand the impact of social habits on the development of OSCC and its influence on the association of HPV16 with OSCC within the OSCC patient cohort, the socio-demographic profile of our OSCC population was collected. Based on our data, the following social habits of patients represented the most common risk factors related to the development of OSCC: tobacco smoking, alcohol drinking and betel quid chewing. However, betel quid chewing remains to be the highest possible risk factor for the development of OSCC among the Malaysian population. In addition, female (67.0%) Indians (49.5%) were the most commonly diagnosed with OSCC when compared to the other two major populations in Malaysia (i.e., Malay and Chinese). This may be related to the predominant personal habits of the female Indian population, such as betel quid chewing [38]. As discussed above, the interaction between gender, race, and HPV infection showed a noticeable impact on the odds of OSCC when both risk factors was present. Whether there is any relationship between these aspects and other social habits with regard to the presence of HPV and the development of OSCC remains unclear. Thus, additional studies are needed to fully understand the influence of these factors.
Conclusion
In summary, we were able to detect the presence of HPV16-specific IgG and IgM antibodies in the sera of OSCC patients, supporting previous reports that HPV16 infection is most likely to be involved in the development of OSCC. Furthermore, our findings indicate that patients with past exposure to HPV infection are at higher risk for OSCC. Importantly, this suggests that HPV antibody detection could be used as a significant indicator of OSCC risk. Therefore, screening for prior HPV infection may help in the prevention and/or early diagnosis of OSCC. Notably, we also confirmed our results by DNA amplification procedures, such as nPCR. However, the high number of control samples with seropositivity for HPV IgG, as well as the overlapping in positivity for IgG and IgM in sera, suggested that antibody detection may be useful for initial screening of a vast number of samples, followed by further comprehensive analyses to validate the results.
Abbreviations
NCR: National Cancer Registry; OSCC: oral squamous cell carcinoma; HPV: human papillomavirus; HR-HPV: high-risk HPV; HNSCC: head and neck squamous cell carcinomas; IgG: immunoglobulin G; IgA: immunoglobulin A; IgM: immunoglobulin M; ELISA: enzyme-linked immunosorbant assay; DNA: Deoxyribonucleic acid; PCR: Polymerase Chain Reaction; nPCR: nested PCR; MOCDTBS: Malaysian Oral Cancer Database & Tissue Bank System; UM: University of Malaya; OCRCC: Oral Cancer and Research Coordinating Center; HRP: horseradish peroxidase; TMB: tetramethylbenzidine; OD: optical density; TBE: Tris-borate-EDTA; RNA: Ribonucleic acid
Acknowledgements
This project was supported by the High Impact Research MoE Grant UM.C/625/1/HIR/MOE/DENT/09 from the Ministry of Education of Malaysia. Samples used in the study were provided by the Oral Cancer Research Coordinating Center (University of Malaya). Jesinda P. Kerishnan was supported by the University of Malaya BrightSpark Scheme. We would also like to thank the following undergraduates for their assistance on some of the experimental work: Ming-Kit Mah and Nurul Ain binti Mohd Fawzi.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lim GCC, Rampal S, Yahaya H. Cancer Incidence in Peninsular Malaysia, 2003-2005: The Third Report of the National Cancer Registry, Malaysia. National Cancer Registry. 2008
2. Walker DM, Boey G, McDonald LA. The pathology of oral cancer. Pathology. 2003;35:376-83
3. Zain RB, Ikeda N, Gupta PC, Warnakulasuriya S, van Wyk CW, Shrestha P. et al. Oral mucosal lesions associated with betel quid, areca nut and tobacco chewing habits: consensus from a workshop held in Kuala Lumpur, Malaysia, November 25-27, 1996. J Oral Pathol Med. 1999;28:1-4
4. Scully C, Bagan J. Oral squamous cell carcinoma: overview of current understanding of aetiopathogenesis and clinical implications. Oral Dis. 2009;15:388-99
5. Ringström E, Peters E, Hasegawa M, Posner M, Liu M, Kelsey KT. Human papillomavirus type 16 and squamous cell carcinoma of the head and neck. Clinical cancer research. 2002;8:3187-92
6. Chen Y, Azman SN, Kerishnan JP, Zain RB, Chen YN, Wong YL. et al. Identification of host-immune response protein candidates in the sera of human oral squamous cell carcinoma patients. PLoS One. 2014;9:e109012
7. Arbyn M, Tommasino M, Depuydt C, Dillner J. Are 20 human papillomavirus types causing cervical cancer? The Journal of pathology. 2014;234:431-5
8. Ono K, Sugahara K, Nomura T, Takano N, Shibahara T, Katakura A. Multiple HPV subtypes infection in Japanese oral squamous cell carcinoma. Journal of Oral and Maxillofacial Surgery, Medicine, and Pathology. 2014;26:128-32
9. Termine N, Giovannelli L, Rodolico V, Matranga D, Pannone G, Campisi G. Biopsy vs. brushing: comparison of two sampling methods for the detection of HPV-DNA in squamous cell carcinoma of the oral cavity. Oral oncology. 2012;48:870-5
10. Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F. et al. A review of human carcinogens—Part B: biological agents. The lancet oncology. 2009;10:321-2
11. McKaig RG, Baric RS, Olshan AF. Hunan papillomavirus and head and neck cancer: Epidemiology and molecular biology. Head and Neck-Journal for the Sciences and Specialties of the Head and Neck. 1998;20:250-65
12. Nelke K, Lysenko L, Leszcyszyn J, Gerber H. Humn papillomavirus and its influence on head and neck cancer predisposition. Postepy Hig Med Dosw. 2013;67:610-6
13. Saini R, Tang TH, Zain RB, Cheong SC, Musa KI, Saini D. et al. Significant association of high-risk human papillomavirus (HPV) but not of p53 polymorphisms with oral squamous cell carcinomas in Malaysia. Journal of cancer research and clinical oncology. 2011;137:311-20
14. Balaram P, Nalinakumari KR, Abraham E, Balan A, Hareendran NK, Bernard HU. et al. Human Papillomaviruses In 91 Oral Cancers From Indian Betel Quid Chewers - High Prevalence And Multiplicity Of Infections. International Journal of Cancer. 1995;61:450-4
15. Sitas F, Urban M, Stein L, Beral V, Ruff P, Hale M. et al. The relationship between anti-HPV-16 IgG seropositivity and cancer of the cervix, anogenital organs, oral cavity and pharynx, oesophagus and prostate in a black South African population. Infect Agent Cancer. 2007;2:6
16. Harro CD, Pang YY, Roden RB, Hildesheim A, Wang Z, Reynolds MJ. et al. Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J Natl Cancer Inst. 2001;93:284-92
17. Ho GY, Studentsov YY, Bierman R, Burk RD. Natural history of human papillomavirus type 16 virus-like particle antibodies in young women. Cancer Epidemiology Biomarkers & Prevention. 2004;13:110-6
18. Petter A, Heim K, Guger M, Ciresa-Ko Nig A, Christensen N, Sarcletti M. et al. Specific serum IgG, IgM and IgA antibodies to human papillomavirus types 6, 11, 16, 18 and 31 virus-like particles in human immunodeficiency virus-seropositive women. J Gen Virol. 2000;81:701-8
19. Syrjanen S, Lodi G, von Bultzingslowen I, Aliko A, Arduino P, Campisi G. et al. Human papillomaviruses in oral carcinoma and oral potentially malignant disorders: a systematic review. Oral Dis. 2011;17(Suppl 1):58-72
20. Zain RB, Athirajan V, Ghani WM, Razak IA, Raja Latifah RJ, Ismail SM. et al. An oral cancer biobank initiative: a platform for multidisciplinary research in a developing country. Cell and tissue banking. 2013;14:45-52
21. Zehbe I, Wilander E. Two consensus primer systems and nested polymerase chain reaction for human papillomavirus detection in cervical biopsies: A study of sensitivity. Human Pathology. 1996;27:812-5
22. Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlee F, Hildesheim A. et al. Improved amplification of genital human papillomaviruses. Journal of Clinical Microbiology. 2000;38:357-61
23. Winder DM, Ball SL, Vaughan K, Hanna N, Woo YL, Franzer JT. et al. Sensitive HPV detection in oropharyngeal cancers. BMC cancer. 2009;9:440
24. Erhart SMM, Rivero ERC, Bazzo ML, Onofre ASC. Comparative evaluation of the GP5+/6+, MY09/11 and PGMY09/11 primer sets for HPV detection by PCR in oral squamous cell carcinomas. Experimental and molecular pathology. 2016;100:13-6
25. de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. The Journal of general virology. 1995;76( Pt 4):1057-62
26. Fuessel Haws AL, He Q, Rady PL, Zhang L, Grady J, Hughes TK. et al. Nested PCR with the PGMY09/11 and GP5(+)/6(+) primer sets improves detection of HPV DNA in cervical samples. Journal of virological methods. 2004;122:87-93
27. van den Brule AJC, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer C, Snijders PJF. GP5+/6+PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. Journal of Clinical Microbiology. 2002;40:779-87
28. Rocha-Zavaleta L, Ambrosio JP, Mora-Garcia Mde L, Cruz-Talonia F, Hernandez-Montes J, Weiss-Steider B. et al. Detection of antibodies against a human papillomavirus (HPV) type 16 peptide that differentiate high-risk from low-risk HPV-associated low-grade squamous intraepithelial lesions. The Journal of general virology. 2004;85:2643-50
29. Kreimer AR, Johansson M, Waterboer T, Kaaks R, Chang-Claude J, Drogen D. et al. Evaluation of Human Papillomavirus Antibodies and Risk of Subsequent Head and Neck Cancer. Journal of Clinical Oncology. 2013;31:2708 -+
30. Wong GR, Ha KO, Himratul-Aznita WH, Yang YH, Wan Mustafa W, Yuen KM. et al. Seropositivity of HPV 16 E6 and E7 and the risk of oral cancer. Oral diseases. 2014
31. Uotila M, Ruoslahti E, Engvall E. Two-site sandwich enzyme immunoassay with monoclonal antibodies to human alpha-fetoprotein. Journal of Immunological Methods. 1981;42:11-5
32. Lim MJ, Foster GJ, Gite S, Ostendorff HP, Narod S, Rothschild KJ. An ELISA-based high throughput protein truncation test for inherited breast cancer. Breast Cancer Res. 2010;12:R78
33. Pintos J, Black MJ, Sadeghi N, Ghadirian P, Zeitouni AG, Viscidi RP. et al. Human papillomavirus infection and oral cancer: A case-control study in Montreal, Canada. Oral oncology. 2008;44:242-50
34. Dillner J. The serological response to papillomaviruses. Seminars in Cancer Biology. 1999;9:423-30
35. Dillner J, Knekt P, Schiller JT, Hakulinen T. Prospective Seroepidemiological Evidence That Human Papillomavirus Type-16 Infection Is A Risk Factor For Esophageal Squamous-Cell Carcinoma. British Medical Journal. 1995;311:1346 -
36. Herrero R, Castellsagué X, Pawlita M, Lissowska J, Kee F, Balaram P. et al. Human papillomavirus and oral cancer: The international agency for research on cancer multicenter study. Journal of the National Cancer Institute. 2003;95:1772-83
37. Mork J, Lie AK, Glattre E, Clark S, Hallmans G, Jellum E. et al. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. New England Journal of Medicine. 2001;344:1125-31
38. Ghani WM, Razak IA, Yang Y-H, Talib NA, Ikeda N, Axell T. et al. Factors affecting commencement and cessation of betel quid chewing behaviour in Malaysian adults. BMC Public Health. 2011;11:82
39. Petti S, Scully C. Determinants of oral cancer at the national level: just a question of smoking and alcohol drinking prevalence? Odontology / the Society of the Nippon Dental University. 2010;98:144-52
40. Neville BW, Day TA. Oral cancer and precancerous lesions. CA: a cancer journal for clinicians. 2002;52:195-215
41. Fakhry C, D'Souza G. Discussing the diagnosis of HPV-OSCC: Common questions and answers. Oral oncology. 2013;49:863-71
42. Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and-unrelated oral squamous cell carcinomas in the United States. Journal of Clinical Oncology. 2008;26:612-9
43. Yen CY, Lu MC, Tzeng CC, Huang JY, Chang HW, Chen RS. et al. Detection of EBV infection and gene expression in oral cancer from patients in Taiwan by microarray analysis. Journal of biomedicine & biotechnology. 2009;2009:904589
44. Yang S-W, Lee Y-S, Chen T-A, Wu C-J, Tsai C-N. Human papillomavirus in oral leukoplakia is no prognostic indicator of malignant transformation. Cancer Epidemiology. 2009;33:118-22
45. Haff LA. Improved Quantitative Pcr Using Nested Primers. Pcr-Methods and Applications. 1994;3:332-7
46. Ostwald C, Muller P, Barten M, Rutsatz K, Sonnenburg M, Milde-Langosch K. et al. Human papillomavirus DNA in oral squamous cell carcinomas and normal mucosa. J Oral Pathol Med. 1994;23:220-5
Author contact
![]() Corresponding author: E-mail:chenyengedu.my; Tel/Fax: (603) 7967 6470
Corresponding author: E-mail:chenyengedu.my; Tel/Fax: (603) 7967 6470

 Global reach, higher impact
Global reach, higher impact