Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(5):386-394. doi:10.7150/ijms.15057 This issue Cite
Research Paper
OCT4 Remodels the Phenotype and Promotes Angiogenesis of HUVECs by Changing the Gene Expression Profile
1. Key Laboratory of Pathobiology, Ministry of Education, Norman Bethune College of Medicine, Jilin University, Changchun, P.R. China.
2. Life Science Research Center, Beihua University, Jilin, P.R. China.
3. The Second Hospital of Jilin University, Changchun, P.R. China.
Received 2016-1-23; Accepted 2016-4-12; Published 2016-4-29
Abstract
It has been shown that forced expression of four mouse stem cell factors (OCT4, Sox2, Klf4, and c-Myc) changed the phenotype of rat endothelial cells to vascular progenitor cells. The present study aimed to explore whether the expression of OCT4 alone might change the phenotype of human umbilical vein endothelial cells (HUVECs) to endothelial progenitor cells and, if so, to examine the possible mechanism involved. A Matrigel-based in vitro angiogenesis assay was used to evaluate the angiogenesis of the cells; the gene expression profile was analyzed by an oligonucleotide probe-based gene array chip and validated by RT-QPCR. The cellular functions of the mRNAs altered by OCT4 were analyzed with Gene Ontology. We found that induced ectopic expression of mouse OCT4 in HUVECs significantly enhanced angiogenesis of the cells, broadly changed the gene expression profile and particularly increased the expression of CD133, CD34, and VEGFR2 (KDR) which are characteristic marker molecules for endothelial progenitor cells (EPCs). Furthermore by analyzing the cellular functions that were targeted by the mRNAs altered by OCT4 we found that stem cell maintenance and cell differentiation were among the top functional response targeted by up-regulated and down-regulated mRNAs upon forced expression of OCT4. These results support the argument that OCT4 remodels the phenotype of HUVECs from endothelial cells to EPCs by up-regulating the genes responsible for stem cell maintenance and down-regulating the genes for cell differentiation.
Keywords: Endothelial Progenitor Cells, Human Umbilical Vein Endothelial Cells, Angiogenesis, Gene Expression, Octamer-binding transcription factor 4.
Introduction
Studies have shown that in adult bone marrow and circulating blood there is a population of cells similar to embryonic angioblasts, known as endothelial progenitor cells (EPCs). These cells are types of stem/progenitor cells with the potential to differentiate into mature endothelial cells and to settle among injured vascular endothelial cells in order to repair damaged blood vessels. In humans, EPCs have been characterized as CD133, CD34, and VEGFR2 (KDR) positive cells [1-4].
The identification of EPCs in adult bone marrow and circulating blood, revised the dogma on adult vascularization from one in which angiogenesis was the only process active in adult vascularization. This earlier concept speculates that circulating endothelial cells (CECs) which had emerged from existing endothelial structures contribute to formation of distant vascular structures. A newer construct proposes that this process, now identified as postnatal vasculogenesis, is a type of adult neovascularization, dependent on bone marrow derived EPCs [1, 5]. In contrast with endothelial cells, EPCs have a much stronger ability to proliferate and to contribute to angiogenesis [6, 7]. Accumulated evidence has shown the importance of EPCs for neovascluraization and vascular remodeling [8, 9]. EPCs have been used in the treatment of vascular diseases [10], promoting reconstruction of ischemic region [11], and have recently played an important role in regeneration medicine [12, 13]. Nonetheless, the limited availability of EPCs is still a bottle neck that restricts their broad application in regenerative medicine.
One of the important potential sources of EPCs is from the differentiation of embryonic stem cells. Studies have demonstrated that endothelial cells (ECs) and smooth muscle cells (SMCs) are both separate cell lineages derived from human embryonic stem cells [14-16]. Human embryonic stem cell-derived EPCs and smooth muscle progenitor cells (SMPCs) are capable of endothelial and smooth muscle cell function. This research has defined the developmental origin and revealed the relationship between these two cell types and provides a complete biological characterization. The discovery that forced expression of the four transcription factors OCT4, Sox2, Klf4, and c-Myc is sufficient to confer a pluripotent state upon the murine and human fibroblast genome, generating induced pluripotent stem cells (iPSCs). These cells have properties similar to embryonic stem cells (ESCs) with regard to their multilineage differentiation potential in vitro and in vivo [17, 18]. The discovery of iPSCs resolved the ethical issues which has plagued the application of ESCs in regenerative medicine. Since then, the rapid progress has been made in the studies on the ways to generate iPSCs from various somatic cells with the defined factors, including skin fibroblasts [18, 19], keratinocytes [20], endothelial cells [21], and blood progenitor cells [22]. For example, Yin L. et al by partially reprogramming rat endothelial cells with the same four transcription factors originally described by Yamanaka [17] forced their expression in rat aorta endothelial cells to successfully generate induced vascular progenitor cells (iVPCs) [23]. These cells remained committed to vascular lineage and could differentiate into vascular ECs and vascular smooth muscle cells (VSMCs) via EPCs and SMPCs in vitro [23]. These cells were demonstrated better in vitro angiogenic potential than native ECs [23].
To decrease the risk of teratoma formation, great efforts have been made to generate iPSCs by decreasing the number of factors used. In this respect, octamer binding transcription factor 4 (OCT4), also known as POU domain, class 5, transcription factor 1 (POU5F1) alone has been successfully used to generate iPSCs from human fetal neural stem cell [24]. OCT4 has also been found to be essential for the maintenance stem-ness of embryonic stem cells [25] and its expression is normally confined to pluripotent cells of embryos [26]. However, research on whether OCT4 alone might induce human EPCs from ECs has not been reported. Based on the evidence described above the present studies were carried out to explore whether forced expression of OCT4 might generate EPCs from HUVECs and, if so, to elucidate the possible mechanism involved.
Materials and Methods
Materials
HUVECs and endothelial cell medium (ECM) were from the ScienCell Research Laboratories (San Diego, USA). Doxycycline (DOX) was purchased from Sigma (St. Louis, USA). Fetal bovine serum (FBS) was from HyClone Inc. (Logan, USA). The Lentiviral Packaging Kit was purchased from Biowit Tech. (Shenzhen, China). The plasmids FUW-M2rtTA and TetO-FUW-OCT4 were from Addgene (Cambridge, USA). In Vitro Angiogenesis Assay Kit was from Millipore (Billerica, USA). Calcein-AM was purchased from Santa Cruz Biotechnology, Inc. (Dallas, USA). PCR primers were synthesized from Sangon Biotec. (Shanghai, China). Trizol Reagent, RT-reaction Kit, and SYBR® Green PCR Master Mix were purchased from TaKaRa Biotec. (Dalian, China).
Cell culture and treatments
The HUVECs were grown in ECM medium containing 5% FBS and 1% endothelial cell growth supplement (ECGS) at 37°C in 5% CO2 and humidified atmosphere. Cells were used for all experiments at passages 2 to 6. For OCT4 induction, the cells were plated in dishes of a 6 cm diameter at a density of 0.5 × 106 cells per dish. After incubating them for 24 hours, the medium was exchanged with fresh medium containing DOX (2 µg/ml) or vehicle and was changed every other day until 7 days when all the cells were harvested.
Transduction of HUVECs
The plasmids FUW-M2rtTA and TetO-FUW-OCT4 were purified with an Endo-Free Plasmid Mini Kit (OMEGA, Norcross, USA). The pseudo-virus packaging was performed by using lentiviral packaging kit according to manufacturer's instruction in 293-T cells. The supernatants were collected at 48h and 72h after transfection and the pseudo-virus were concentrated by high-speed centrifugation (50000g for 2 hour at 4°C). HUVECs were transduced by using the pseudo-virus and polybrene (4μg/ml) for 24 hours. The medium was changed on the second day.
RNA purification and RT-QPCR
Total RNA from the cells was purified with a TRIzol Reagent following the manufacturer's instruction. The purity and quantity of the RNA was measured with spectrophotometer and the quality of RNA was further monitored by agarose gel electrophoresis. After treatment with RNase-free DNase I, RNA was subjected to reverse transcription with a RT-reaction Kit. The cDNA product was amplified and quantified with 7300 Real-time PCR system (Applied Biosystems) in a 25 μl reaction volume using SYBR® Green PCR Master Mix. The primer sets used for PCR amplification are shown in Table 1. The thermal cycling program consisted of 2 min at 50°C, 10 min at 95°C, followed by 40 cycles for 15 sec at 95°C and 1 min at 60°C. After amplification, a melting curve was generated and data analysis was performed by using Dissociation Curves 1.0 software (Applied Biosystems). The normalized value was given by the ratio of mRNA of the target gene to mRNA of the reference gene (RPL13A) in each sample. Fold activation was given by the ratio of the normalized values of the cells incubated with (+DOX) to that without (-DOX) DOX.
Primer sets used for RT-QPCR.
| Gene | Primer sets | Sequences | GenBank |
|---|---|---|---|
| hRPL13A | Forward | 5'-CGAGGTTGGCTGGAAGTACC-3' | NM_012423 |
| Reverse | 5'-CTTCTCGGCCTGTTTCCGTAG-3' | ||
| mOCT4 | Forward | 5'-CAGCCAGACCACCATCTGTC-3' | NM_013633 |
| Reverse | 5'-GTCTCCGATTTGCATATCTCCTG-3' | ||
| hOCT4 | Forward | 5'-GGGAGATTGATAACTGGTGTGTT-3' | NM_203289 |
| Reverse | 5'-GTGTATATCCCAGGGTGATCCTC-3' | ||
| hKDR | Forward | 5'-GTGATCGGAAATGACACTGGAG-3' | NM_002253 |
| Reverse | 5'-CATGTTGGTCACTAACAGAAGCA-3' | ||
| hCD34 | Forward | 5'-CTACAACACCTAGTACCCTTGGA-3' | NM_001773 |
| Reverse | 5'-GGTGAACACTGTGCTGATTACA-3' | ||
| hCD133 | Forward | 5'-CCTCATGGTTGGAGTTGGAT-3' | NM_006017 |
| Reverse | 5'-TTCCACATTTGCACCAAAGA-3' | ||
| hAVIL | Forward | 5'-ACAACGACCCTGGGATCATTG-3' | NM_006576 |
| Reverse | 5'-GTCGAGAGGATGACGTAGCAG-3' | ||
| hS100A4 | Forward | 5'-GATGAGCAACTTGGACAGCAA-3' | NM_002961 |
| Reverse | 5'-CTGGGCTGCTTATCTGGGAAG-3' | ||
| hSLC12A3 | Forward | 5'-CTCCACCAATGGCAAGGTCAA-3' | NM_000339 |
| Reverse | 5'-GGATGTCGTTAATGGGGTCCA-3' | ||
| hS100P | Forward | 5'-AAGGATGCCGTGGATAAATTGC-3' | NM_005980 |
| Reverse | 5'-ACACGATGAACTCACTGAAGTC-3' | ||
| hFOLR1 | Forward | 5'-GCTCAGCGGATGACAACACA-3' | NM_000802 |
| Reverse | 5'-CCTGGCCCATGCAATCCTT-3' | ||
| hIQCF1 | Forward | 5'-CAGCCCCAAAAGACGAAGGAA-3' | NM_152397 |
| Reverse | 5'-GCTCCTAAGGACAAATGGGTTG-3' | ||
| hCD31 | Forward | 5'-AACAGTGTTGACATGAAGAGCC-3' | NM_000442 |
| Reverse | 5'-TGTAAAACAGCACGTCATCCTT-3' | ||
| hVE-Cadherin | Forward | 5'-TTGGAACCAGATGCACATTGAT-3' | NM_001795 |
| Reverse | 5'-TCTTGCGACTCACGCTTGAC-3' | ||
| hvW-Factor | Forward | 5'-CCGATGCAGCCTTTTCGGA-3' | NM_000552 |
| Reverse | 5'-TCCCCAAGATACACGGAGAGG-3' |
In vitro angiogenesis assay
The angiogenesis of the cells was evaluated by a Matrigel in vitro angiogenesis assay technique [27, 28]. Briefly, 100μl stock solution of Matrigel was added to each well in 48-well plates and kept at 37°C for 30 min in order to form the Matrigel. Cell suspensions containing 3×104 cells in 100μl of ECM were seeded on the Matrigel of each well, and incubated for 6 hours. Then Calcein-AM (0.1 mM) was directly added to each well for 20 min at 37°C to stain the cells and imaged under a phase contrast microscope with an excitation wavelength of 490 nm and an emission wavelength of 515 nm. For quantification, the values for the pattern recognition, branch point and total capillary tube length were determined following the manufacturer's guidelines (ECM625; Millipore). ImageJ software was used in the first instance prior to double-checking by an independent assessor. 5 random microscopic (×100) fields per well were included and the data are expressed as Mean±SD of 5 samples.
Gene expression profiling analysis
Whole-genome expression arrays were performed by using Roche NimbleGen chips (KangChen, Shanghai, China), an oligonucleotide-probe-based gene array chip containing 45,033 transcripts, which provides a comprehensive coverage of the whole human genome. Total RNA from each sample was isolated and quantified by the NanoDrop ND-1000. The integrity of RNA was assessed by standard denaturing agarose gel electrophoresis. Total RNA was used to synthesize cDNA by reverse transcription reaction, subsequently, which was labeled with a NimbleGen one-color DNA labeling kit, and then Hybridized using NimbleGen Hybridization System following the manufacturer's instruction. The chip was washed, and scanned with Axon GenePix 4000B. Following normalization, all files of gene expression level were imported into Agilent GeneSpring GX software (version 11.5) for further analysis. Genes that have values greater than or equal to lower cut-off: 100.0 were chosen for differentially expressed gene screening. After data filtering, scatter plot analysis was performed to assess gene expression data. The values of X and Y axes in the Scatter-Plot are the averaged normalized signal values of each group (log2 scaled). The green lines are Fold Change Lines (The default fold change value given was 2.0).
Bioinformatics analysis
Gene Ontology (GO) [29] is a functional analysis to interrogate the possible functions associated with the differentially expressed genes. Following data filtering based on the statistical standard, differentially expressed genes were included in the analysis. The p-value denotes the significance of GO Term enrichment in the differentially expressed gene list. The lower the p-value, the more significant the GO term is. FDR stands for the false discovery rate of the GO item. The lower the FDR value, the less the false discovery rate of the GO item is [29].
Statistical analysis
All calculations and statistical analyses were performed by using GraphPad Prism 5.0 software (San Diego, CA, USA). T test was used to analyze the significance of any differences between two groups. The statistical significance was defined as p<0.05.
Results
Induced expression of OCT4 in HUVECs
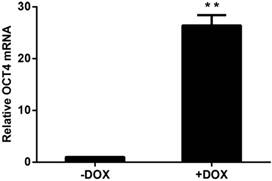
HUVECs that were lentivirally transduced with the Tet-on controlled OCT4 expression vector were incubated in the presence of DOX or vehicle to induce OCT4 expression in the cells. OCT4 mRNA was analyzed by RT-QPCR. The results are shown in Fig. 1. OCT4 mRNA was increased by more than 20 fold in the cells treated with DOX, when compared with the cells not exposed to DOX. These results indicate that the cellular model for DOX induced OCT4 expression was established.
Induced expression of OCT4 in HUVECs by DOX. The mRNAs were analyzed by RT-QPCR and the amount of OCT4 mRNA was normalized to internal standard RPL13A mRNA. Relative fold was calculated based on the ratio of the normalized values of the cells incubated with (+DOX) to that without (-DOX) DOX. The data are expressed as Mean±SD, N=3, ** P<0.01 versus that of -DOX cells.
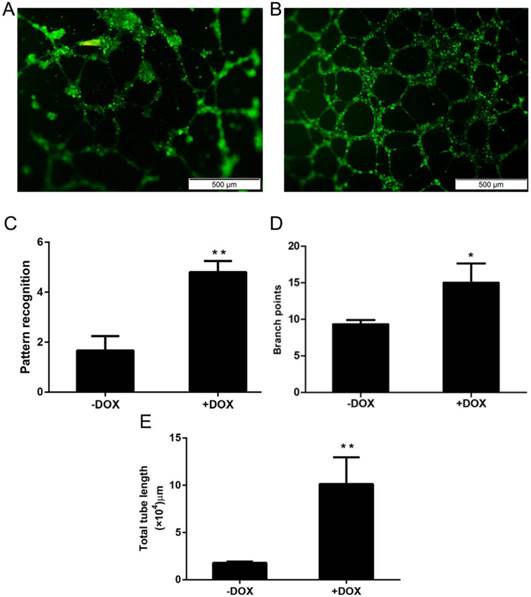
OCT4 enhanced the angiogenesis of HUVECs
Angiogenesis is the major function of vascular endothelial cells (ECs) and their precursor, endothelial progenitor cells (EPCs), which have a potential application for cell therapy because they have a much stronger ability for angiogenesis, when compared with mature ECs [6, 7]. To determine whether OCT4 can enhance the angiogenesis of HUVECs or not, an in vitro angiogenesis assay system was used to evaluate the changes of in vitro angiogenesis of the cells, based on the formation of tubular networks. The angiogenesis assay was performed for HUVECs which were lentivirally transduced with the Tet-on controlled OCT4 expression vector and incubated with or without Tet-on inducer DOX. The results are shown in Fig. 2, in which A and B are representative microscopic appearances and C-E where the results are statistically analyzed. Clearly, more tubular networks were formed in the cells induced with DOX (+DOX) and a lower percentage of network formations was found in un-induced cells (-DOX). These results indicate that forced expression of OCT4 enhanced angiogenesis of HUVECs, suggesting that OCT4 might remodel the phenotype of the cells from ECs to EPCs.
OCT4 altered gene expression profiles in HUVECs
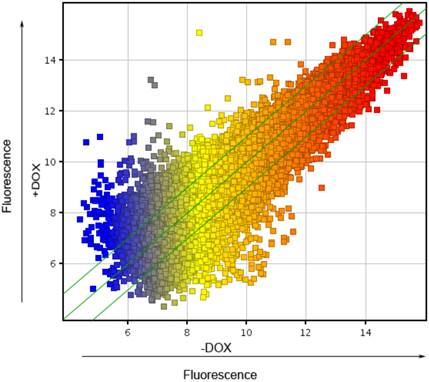
To explore the molecular mechanism, underlying the finding that forced expression of OCT4 enhanced angiogenesis of HUVECs, whole genome expression arrays were carried out in HUVECs lentivirally transduced with the Tet-on controlled OCT4 expression vector and incubated in the presence of DOX or vehicle. The Scatter-Plot analysis of the changes in the gene expression profile was made to show the global change of gene expression profile in HUVECs induced by DOX. This is shown in Fig. 3. Each point on the scatter plot represents the expression level of an individual mRNA, as determined by units of fluorescence intensity. The values of X and Y axes of each point in the Scatter-Plot represent the levels of respective mRNA of the cells treated with (+DOX in Fig. 3) and without DOX (-DOX in Fig. 3), respectively. The default fold change value given is 2.0 (within the range of green lines). The points above the top and below the bottom green lines represent the genes whose mRNA changed more than 2 fold between two groups of cells. The numbers of genes whose expression was up-regulated were 530 genes over 2 fold, 44 genes over 5 fold and 19 genes over 10 fold. In contrast, the numbers of genes whose expression was down-regulated were 502 genes over 2 fold, 68 genes over 5 fold and 20 genes over 10 folds. The results indicate that the expression of OCT4 broadly changed the gene expression profile of HUVECs.
OCT4 enhanced angiogenesis of HUVECs. The angiogenesis of the cells was evaluated by an in vitro angiogenesis assay kit, as described in the method section. A and B are representative microscopic photographs of uninduced and induced cells. C, D and E are statistically analyzed results. N=5, *P<0.05 and **P<0.01 versus that of -DOX cells.
Scatter plot analysis of genes regulated by OCT4 in HUVECs. A scatter chart for the normalized mRNA expression data, determined by using Microarray analysis, was made to show the general change of gene expression profile in HUVECs induced by DOX. +DOX and -DOX represent the mRNA values in the cells incubated with and without DOX, respectively. Each point on the scatter plot represents the expression level of an individual mRNA, as determined by units of fluorescence intensity. The default fold change value given is 2.0 (within the range of green lines). The points above the top and below the bottom green lines represent the mRNAs whose values changed more than 2 fold between two groups of cells. N=3.
To focus on the genes whose expression was extremely altered by OCT4, the 20 most up-regulated and down-regulated mRNAs were identified and are shown in Table 2 and Table 3, respectively. Table 2 lists the 20 mRNAs that were most up-regulated by induction of OCT4 in HUVECs. Most of these mRNAs were increased by more than 10 fold in magnitude (with the exception of only one mRNA which was increased by more than 9 fold). Interestingly, OCT4 mRNAs (POU5F1 in Table 2) and the other three mRNAs (CD133, CD34, and VEGFR2) were among them. OCT4 is a transcription factor of stem cells and maintains the stem-ness of the cells by a series of complex regulated positive feedback networks. Increased expression of human OCT4 mRNAs indicates that the induction of ectopic mouse OCT4 can initiate the transcription of the endogenous human OCT4 gene, whose transcript is undetectable in HUVECs. CD133, CD34, and VEGFR2 have been accepted, as three molecular markers of human EPCs [1-4]. The results suggest that the forced expression of OCT4 can remodel the phenotype of HUVECs from ECs to EPCs. Table 3 lists the 20 mRNAs that were most down-regulated by OCT4 in HUVECs. Most of them, with the exception of two mRNAs, were decreased more than 5 fold in the magnitude.
To validate the alteration of mRNAs by OCT4 in HUVECs, expression of thirteen of the mRNAs was further confirmed by RT-QPCR. These were chosen based on five of the mRNAs which were up-regulated over tenfold: AVIL (85.2), OCT4 (14.1), KDR (15.8), CD34 (13.2), CD133 (11.0); and eight of down-regulated: S100A4 (0.14), SLC12A3 (0.16), S100P (0.08), FOLR1 (0.11), IQCF1 (0.24), CD31 (0.36), VE-Cadherin (0.15) and vW-Factor (0.60). These all demonstrated either more than tenfold changes in expression or are molecular marker of HUVECs. As shown in Table 4, all of the 13 mRNAs were analyzed showed the same pattern of change in expression by the two techniques. Our results indicate the reliability of the Microarray results. Particularly, we want to point out that human OCT4 and three molecular markers for EPCs, CD133, CD34, and VEGFR2 (KDR), were among the mRNAs whose expressions were highly increased upon treatment with DOX determined by two techniques. The results support above conclusion that phenotypic remodeling of the cells from ECs to EPCs might occur by forced expression of OCT4. The two methods also showed that the mRNAs CD31, vascular endothelial cadherin (VE-cadherin) and von Willebrand Factor (vW-Factor), which are the molecular markers for mature endothelial cells, but not expressed in their progenitor cells, were decreased upon DOX treatment. This result provided additional evidence to support our conclusion.
Top 20 genes up-regulated by OCT4 in HUVECs.
| Genes | Fold Increase | Gene Name | GenBank | |
|---|---|---|---|---|
| 1 | KRT10 | 97.0 ± 11.79** | keratin 10 (epidermolytic hyperkeratosis; keratosis palmaris et plantaris) | NM_000421 |
| 2 | AVIL | 85.2 ± 20.39** | advillin | NM_006576 |
| 3 | KRT20 | 31.0 ± 6.12** | keratin 20 | NM_019010 |
| 4 | FMN1 | 30.5 ± 13.67** | formin 1 | BC103692 |
| 5 | EHF | 29.9 ± 18.73** | ets homologous factor | AF124438 |
| 6 | PRND | 27.2 ± 5.97** | prion protein 2 (dublet) | NM_012409 |
| 7 | OTOR | 22.4 ± 11.89** | otoraplin | NM_020157 |
| 8 | KDR | 15.9 ± 1.57** | kinase insert domain receptor (a type III receptor tyrosine kinase) | NM_002253 |
| 9 | FLJ46906 | 15.5 ± 3.65** | hypothetical gene supported by AK128874; BC071813 | XM_928441 |
| 10 | GPNMB | 14.2 ± 3.70** | glycoprotein (transmembrane) nmb | BC011595 |
| 11 | POU5F1 | 14.1 ± 0.82** | POU domain, class 5, transcription factor 1 | Z11898 |
| 12 | CD34 | 13.2 ± 1.82** | CD34 molecule | BX538076 |
| 13 | FLJ37464 | 13.0 ± 1.55** | hypothetical protein FLJ37464 | AK094783 |
| 14 | KCNMA1 | 11.7 ± 1.05** | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | NM_002247 |
| 15 | RNF36 | 11.4 ± 1.18** | ring finger protein 36 | NM_080745 |
| 16 | CLDN11 | 11.0 ± 5.56** | claudin 11 (oligodendrocyte transmembrane protein) | BC013577 |
| 17 | CD133 | 11.0 ± 2.86** | prominin 1 | NM_006017 |
| 18 | HS6ST3 | 11.0 ± 8.52** | heparan sulfate 6-O-sulfotransferase 3 | XM_931159 |
| 19 | CDH10 | 10.3 ± 2.35** | cadherin 10, type 2 (T2-cadherin) | NM_006727 |
| 20 | SUSD2 | 9.3 ± 1.83** | sushi domain containing 2 | NM_019601 |
The mRNAs data (fluorescence) determined by Microarray assay were normalized through the Roche NimbleScan software. Relative fold values were calculated based on the ratio of the cells incubated with DOX to that without DOX. 20 most up-regulated genes are listed. Each value is the Mean ± SD from triplicate samples. **p<0.01, versus the -DOX cells.
Top 20 genes down-regulated by OCT4 in HUVECs.
| Genes | Fold Decrease | Gene Name | GenBank | |
|---|---|---|---|---|
| 1 | LOC645009 | 0.04 ± 0.02** | similar to GAGE-4 protein (G antigen 4) | BC081536 |
| 2 | GAGE5 | 0.14 ± 0.15** | G antigen 5 | NM_001475 |
| 3 | S100A4 | 0.14 ± 0.06** | S100 calcium binding protein A4 (calcium protein, calvasculin, metastasin, murine placental homolog) | NM_002961 |
| 4 | SLC12A3 | 0.16 ± 0.01** | solute carrier family 12 (sodium/chloride transporters), member 3 | NM_000339 |
| 5 | S100P | 0.08 ± 0.04** | S100 calcium binding protein P | BC006819 |
| 6 | KRTHB1 | 0.05 ± 0.01** | keratin, hair, basic, 1 | NM_002281 |
| 7 | FRMD3 | 0.19 ± 0.20** | FERM domain containing 3 | AY137774 |
| 8 | WISP2 | 0.08 ± 0.04** | WNT1 inducible signaling pathway protein 2 | BC058074 |
| 9 | GAGE8 | 0.10 ± 0.07** | G antigen 8 | NM_012196 |
| 10 | FOLR1 | 0.11 ± 0.09** | folate receptor 1 (adult) | NM_000802 |
| 11 | IQCF1 | 0.24 ± 0.26** | IQ motif containing F1 | NM_152397 |
| 12 | LPAL2 | 0.10 ± 0.05** | lipoprotein, Lp(a)-like 2 | AK125910 |
| 13 | FOLR3 | 0.38 ± 0.44** | folate receptor 3 (gamma) | U08470 |
| 14 | SLC16A6 | 0.17 ± 0.16** | solute carrier family 16 (monocarboxylic acid transporters), member 6 | NM_004694 |
| 15 | A2M | 0.10 ± 0.05** | alpha-2-macroglobulin | NM_000014 |
| 16 | DKFZP686 A01247 | 0.06 ± 0.01** | hypothetical protein | NM_014988 |
| 17 | SNCG | 0.12 ± 0.07** | synuclein, gamma (breast cancer-specific protein 1) | NM_003087 |
| 18 | SVEP1 | 0.14 ± 0.10** | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | XM_933024 |
| 19 | FAM46A | 0.13 ± 0.10** | family with sequence similarity 46, member A | NM_017633 |
| 20 | GDA | 0.13 ± 0.03** | guanine deaminase | NM_004293 |
The mRNAs data (fluorescence) determined by using Microarray assay were normalized through the Roche NimbleScan software. Relative fold expression was calculated based on the ratio of the cells incubated with DOX to that without DOX. 20 most down-regulated genes are listed. Each value is the Mean ± SD from triplicate samples. **p<0.01, versus the -DOX cells.
Validation of the mRNAs expression by RT-QPCR.
| mRNAs | Folds (+DOX/-DOX) | |
|---|---|---|
| MicroArray | RT-QPCR | |
| AVIL | 85.2 ± 20.39** | 12.2 ± 2.64** |
| OCT4 | 14.1 ± 0.82** | 26.4 ± 1.98** |
| KDR | 15.8 ± 1.57** | 7.9 ± 2.20** |
| CD34 | 13.2 ± 1.82** | 10.8 ± 2.22** |
| CD133 | 11.0 ± 2.86** | 8.1 ± 1.47** |
| S100A4 | 0.14 ± 0.06** | 0.14 ± 0.25** |
| SLC12A3 | 0.16 ± 0.01** | 0.23 ± 0.11** |
| S100P | 0.08 ± 0.04** | 0.08 ± 0.04** |
| FOLR1 | 0.11 ± 0.09** | 0.11 ± 0.20** |
| IQCF1 | 0.24 ± 0.26** | 0.12 ± 0.06** |
| CD31 | 0.36 ± 0.24** | 0.12 ± 0.02** |
| VE-Cadherin | 0.15 ± 0.05** | 0.27 ± 0.05** |
| vW-Factor | 0.60 ± 0.17** | 0.37 ± 0.19** |
The mRNAs determined by Microarray assay and RT-QPCR were normalized through the Roche NimbleScan software and by the internal standard RPL13A mRNA, respectively. Relative fold values in expression were calculated based on the ratio of the cells incubated in the presence (+DOX ) to that of the cells in the absence (-DOX ) of DOX. Each value is the Mean ± SD from triplicate samples. **p<0.01, versus the -DOX cells.
Molecular functions of genes regulated by OCT4.
| Up-regulated by OCT4 | p-value* | FDR# |
|---|---|---|
| immune response | 1.3379E-06 | 0.00595899 |
| apoptotic signaling pathway | 7.4837E-06 | 0.007612657 |
| defense response | 2.0037E-05 | 0.044623077 |
| response to stress | 2.2132E-05 | 0.010952796 |
| positive regulation of cellular process | 3.7134E-05 | 0.015073193 |
| multi-organism process | 4.0214E-05 | 0.198188031 |
| response to biotic stimulus | 4.6818E-05 | 0.443289936 |
| stem cell maintenance | 5.0928E-05 | 0.443289936 |
| multicellular organismal signaling | 5.7531E-05 | 0.458326667 |
| positive regulation of biological process | 6.3142E-05 | 0.015624031 |
| Down-regulated by OCT4 | ||
| response to stimulus | 7.1105E-11 | 1.5835E-07 |
| cell differentiation | 2.9436E-09 | 4.15823E-06 |
| system development | 4.2726E-09 | 4.15823E-06 |
| multicellular organismal development | 5.6016E-09 | 4.15823E-06 |
| anatomical structure development | 6.1858E-08 | 2.41044E-05 |
| cell adhesion | 2.8368E-07 | 8.5696E-05 |
| biological adhesion | 3.1936E-07 | 8.89007E-05 |
| cell-cell signaling | 5.425E-07 | 0.000142136 |
| locomotion | 3.9781E-06 | 0.000610987 |
| tissue development | 1.6513E-05 | 0.002228712 |
Gene Ontology (GO) analysis was carried out on the differentially expressed mRNAs determined by Microarray assay to explore their molecular functions. p-value* stands for the significance testing value of the GO item and FDR# stands for the false discovery rate of the GO item.
Cellular functions of the mRNAs regulated by OCT4 in HUVECs
The cellular functions that were targeted by the mRNAs altered by OCT4 were analyzed with Gene Ontology (GO) [29]. Interestingly, stem cell maintenance and cell differentiation were among the top functions targeted by up-regulated and down-regulated mRNAs upon OCT4 treatment, respectively, as shown in Table 5. These results support the argument that OCT4 remodels the phenotype of HUVECs from ECs to EPCs via up-regulating the genes responsible for stem cell maintenance and down-regulating the genes for cell differentiation. Other biological categories of statistical significance targeted by up-regulated mRNAs upon the expression of OCT4 include the apoptotic signaling pathway, defense response, and positive regulation of cellular and biological process. The biological categories targeted by down-regulated mRNAs include the development related (tissue development; system development; and anatomical structure development), cell adhesion and cell-cell signaling. These results gave an additive evidence to support the hypothesis that phenotypic remodeling of the cells from ECs to EPCs might occur by forced expression of OCT4.
Discussion
The limited availability of qualified EPCs is a major concern in regenerative medicine. In the present study, we found that forced expression of OCT4 in HUVECs significantly enhanced the in vitro angiogenesis of the cells. It has been reported that EPCs have much better ability for in vitro angiogenesis than ECs [6, 7]. Based on the major role of OCT4 in maintaining stem-ness of embryonic stem cells [25] and inducing generation of iPSCs from human fetal neural stem cell [24] these results suggest that OCT4 can remodel the phenotype of HUVECs from ECs to EPCs.
Gene expression profile determines the characteristic phenotype and the function of the cells. In the present study, we found that forced expression of ectopic mouse OCT4 broadly changed the gene expression profile of HUVECs. Particularly, it increased the expression of endogenous human OCT4 gene which is a transcription factor of stem cells and plays an important role in keeping the stem-ness of the cells. Its transcript is undetectable in differentiated HUVECs [30, 31], suggesting that phenotypic remodeling of the cells from ECs to EPCs might occur by forced expression of OCT4 alone in HUVECs. Interestingly, the expression of CD133, CD34, and VEGFR2 which have been accepted as characteristic molecular markers for human EPCs [1-4] significantly increased by the forced expression of OCT4 alone in the HUVECs. The expression of CD31, VE-cadherin and vW-Factor, which are the molecular markers for mature ECs, but not expressed in their progenitor cells [32, 33], were significantly decreased upon the expression of OCT4. This result supports the argument that forced expression of OCT4 alone remodels the phenotype of HUVECs from ECs to EPCs.
To explore the molecular mechanism on how the expression of OCT4 affects angiogenesis of HUVECs, our special attention has been paid to the genes, whose upregulation might be involved in angiogenesis, upregulated by OCT4 found in this study. In addition VEGFR2 (KDR in Table 2), whose expression was upregulated about 16 times, the expression of VEGFA, bFGF, eNOS, IL1B and IL6 was also increased significantly upon forced expression of OCT4, although they are not among the top 20 genes. Particularly, VEGFA is among them. It has been reported that VEGF signalling through VEGFR2 is the major angiogenic pathway, and blockage of VEGF/VEGFR2 signalling is the first anti-angiogenic strategy for cancer therapy [34]. It seems to us that increased angiogenesis of HUVECs upon OCT4 expression might be attributed to the upregulation of VEGFA and VEGFR2, although further study is required for the exact mechanism involved. In our knowledge, the upregulation of VEGF/VEGFR2 by OCT4 has not been reported in the cells.
Furthermore, by analyzing the cellular functions targeted by the altered mRNAs, stem cell maintenance and cell differentiation were among the top categories targeted by up-regulated and down-regulated mRNAs following forced expression of OCT4. These results further support the argument that OCT4 can remodel the phenotype of HUVECs from ECs to EPCs via up-regulating the expression of genes responsible for stem cell maintenance and down-regulating the expression of genes for cell differentiation. The results provide additive evidence supporting the hypothesis that the forced expression OCT4 alone in HUVECs might remodel the phenotypes of the cells from ECs to EPCs by broadly changing their gene expression profile. Taken together the results indicate that OCT4 can remodel the phenotype of HUVECs from ECs to EPCs. To our knowledge, this study provides the first evidence indicating that OCT4 alone remodels the phenotype and promotes angiogenesis of HUVECs by changing the gene expression profile. Theoretically, these findings provide more insights on the role of OCT4 in keeping progenitor state of endothelial cells. Practically, the present study might provide an efficient way to generate adequate numbers of qualified EPCs for regenerative medicine.
Conclusions
In the present study, we found that forced expression of mouse OCT4 in HUVECs remodels the phenotype of the cells from ECs to EPCs. This conclusion was supported by the following evidence: firstly forced expression of mouse OCT4 enhanced in vitro angiogenesis of the cells (EPCs have stronger in vitro angiogenesis ability than ECs); it up-regulated the expression of CD133, CD34, and VEGFR2 (they are characteristic molecular markers for human EPCs) and down-regulated CD31, VE-cadherin and vW-Factor (they are the molecular markers for mature ECs, but not expressed in their progenitor cells). Further by analyzing the cellular functions targeted by the mRNAs altered upon OCT4 expression we found that functions associated with stem cell maintenance were targeted by up-regulated mRNAs and cell differentiation functions were targeted by down-regulated mRNAs.
Acknowledgements
This study was supported in part by the National Natural Science Foundation of China (Grants: NSFC No. 21277057) and National Science Foundation of Jilin Province (No. 20130624003JC). We would like to express our great appreciation to Professor F. William Orr from the University of Manitoba in Canada for his great help in revising the manuscript and to Professor Rudolf Jaenisch from the Whitehead Institute for Biomedical Research in USA for his great help in contributing the plasmids FUW-M2rtTA and TetO-FUW-OCT4 to Addgene.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Rafii S, Lyden D. Therapeutic stem and progenitor cell transplantation for organ vascularization and regeneration. Nature medicine. 2003;9:702-12
2. Iwami Y, Masuda H, Asahara T. Endothelial progenitor cells: past, state of the art, and future. Journal of cellular and molecular medicine. 2004;8:488-97
3. Peichev M, Naiyer AJ, Pereira D, Zhu Z, Lane WJ, Williams M. et al. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95:952-8
4. Caiado F, Dias S. Endothelial progenitor cells and integrins: adhesive needs. Fibrogenesis Tissue Repair. 2012:5
5. Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T. et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964-6
6. Yoon C-H, Hur J, Park K-W, Kim J-H, Lee C-S, Oh I-Y. et al. Synergistic neovascularization by mixed transplantation of early endothelial progenitor cells and late outgrowth endothelial cells the role of angiogenic cytokines and matrix metalloproteinases. Circulation. 2005;112:1618-27
7. Sieveking DP, Buckle A, Celermajer DS, Ng MK. Strikingly different angiogenic properties of endothelial progenitor cell subpopulations: insights from a novel human angiogenesis assay. Journal of the American College of Cardiology. 2008;51:660-8
8. Calzi SL, Neu MB, Shaw LC, Kielczewski JL, Moldovan NI, Grant MB. EPCs and pathological angiogenesis: when good cells go bad. Microvascular research. 2010;79:207-16
9. Hristov M, Weber C. Endothelial progenitor cells in vascular repair and remodeling. Pharmacological Research. 2008;58:148-51
10. Schmidt DE, Manca M, Hoefer IE. Circulating endothelial cells in coronary artery disease and acute coronary syndrome. Trends in cardiovascular medicine. 2015
11. Peplow PV. Growth factor-and cytokine-stimulated endothelial progenitor cells in post-ischemic cerebral neovascularization. Neural regeneration research. 2014;9:1425
12. Foster WS, Suen CM, Stewart DJ. Regenerative cell and tissue-based therapies for pulmonary arterial hypertension. Canadian Journal of Cardiology. 2014;30:1350-60
13. Wei L, Zhu W, Xia L, Yang Y, Liu H, Shen J. et al. Therapeutic effect of eNOS-transfected endothelial progenitor cells on hemodynamic pulmonary arterial hypertension. Hypertension Research. 2013;36:414-21
14. Ferreira LS, Gerecht S, Shieh HF, Watson N, Rupnick MA, Dallabrida SM. et al. Vascular progenitor cells isolated from human embryonic stem cells give rise to endothelial and smooth muscle-like cells and form vascular networks in vivo. Circulation research. 2007;101:286-94
15. Hill KL, Obrtlikova P, Alvarez DF, King JA, Keirstead SA, Allred JR. et al. Human embryonic stem cell-derived vascular progenitor cells capable of endothelial and smooth muscle cell function. Exp Hematol. 2010;38:246-57 e1
16. Glaser DE, Gower RM, Lauer NE, Tam K, Blancas AA, Shih AJ. et al. Functional characterization of embryonic stem cell-derived endothelial cells. J Vasc Res. 2011;48:415-28
17. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. cell. 2006;126:663-76
18. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. cell. 2007;131:861-72
19. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-20
20. Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F. et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature biotechnology. 2008;26:1276-84
21. Lagarkova MA, Shutova MV, Bogomazova AN, Vassina EM, Glazov EA, Zhang P. et al. Induction of pluripotency in human endothelial cells resets epigenetic profile on genome scale. cell cycle. 2010;9:937-46
22. Loh Y-H, Agarwal S, Park I-H, Urbach A, Huo H, Heffner GC. et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476-9
23. Yin L, Ohanyan V, Pung YF, DeLucia A, Bailey E, Enrick M. et al. Induction of vascular progenitor cells from endothelial cells stimulates coronary collateral growth. Circulation research. 2012;110:241-52
24. Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H. et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649-53
25. Zeineddine D, Hammoud AA, Mortada M, Boeuf H. The Oct4 protein: more than a magic stemness marker. American journal of stem cells. 2014;3:74
26. Wu G, Schöler HR. Role of Oct4 in the early embryo development. Cell Regeneration. 2014;3:7
27. Salani D, Taraboletti G, Rosano L, Di Castro V, Borsotti P, Giavazzi R. et al. Endothelin-1 induces an angiogenic phenotype in cultured endothelial cells and stimulates neovascularization in vivo. Am J Pathol. 2000;157:1703-11
28. Amini AR, Laurencin CT, Nukavarapu SP. Differential analysis of peripheral blood- and bone marrow-derived endothelial progenitor cells for enhanced vascularization in bone tissue engineering. J Orthop Res. 2012;30:1507-15
29. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-9
30. Yu KR, Yang SR, Jung JW, Kim H, Ko K, Han DW. et al. CD49f enhances multipotency and maintains stemness through the direct regulation of OCT4 and SOX2. Stem cells. 2012;30:876-87
31. Panopoulos AD, Ruiz S, Yi F, Herrerías A, Batchelder EM, Izpisua Belmonte J. Rapid and highly efficient generation of induced pluripotent stem cells from human umbilical vein endothelial cells. PloS one. 2011;6:e19743
32. Barber CL, Iruela-Arispe ML. The ever-elusive endothelial progenitor cell: identities, functions and clinical implications. Pediatric research. 2006;59:26R-32R
33. Medina RJ, O'Neill CL, Sweeney M, Guduric-Fuchs J, Gardiner TA, Simpson DA. et al. Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities. BMC medical genomics. 2010;3:18
34. Lohela M, Bry M, Tammela T, Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol. 2009;21:154-65
Author contact
![]() Corresponding authors: Dr. Ronggui Li, The Key Laboratory of Pathobiology, Ministry of Education, Norman Bethune College of Medicine, Jilin University, Changchun, 130021, P.R. China. Tel.: 86-431 85619481; Fax: 86-431-85619469; E-mail: lirgedu.cn and Dr. Xin Sun, Life Science Research Center, Beihua University, Jilin, 132013, P.R. China. Tel.: 86-432-64608351; E-mail: sunxinbhcom.
Corresponding authors: Dr. Ronggui Li, The Key Laboratory of Pathobiology, Ministry of Education, Norman Bethune College of Medicine, Jilin University, Changchun, 130021, P.R. China. Tel.: 86-431 85619481; Fax: 86-431-85619469; E-mail: lirgedu.cn and Dr. Xin Sun, Life Science Research Center, Beihua University, Jilin, 132013, P.R. China. Tel.: 86-432-64608351; E-mail: sunxinbhcom.

 Global reach, higher impact
Global reach, higher impact