Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(5):330-339. doi:10.7150/ijms.14341 This issue Cite
Research Paper
Secretion of N- and O-linked Glycoproteins from 4T1 Murine Mammary Carcinoma Cells
1. Department of Oral Biology & Biomedical Sciences, Faculty of Dentistry, University of Malaya, Kuala Lumpur 50603, Malaysia
2. Institute for Research in Molecular Medicine (INFORMM), Universiti Sains Malaysia, Penang 11800, Malaysia
3. Institute of Nano Electronic Engineering (INEE), Universiti Malaysia Perlis, 01000 Kangar, Perlis, Malaysia
4. School of Bioprocess Engineering, Universiti Malaysia Perlis, 02600 Arau, Perlis, Malaysia
5. Department of Molecular Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur 50603, Malaysia
6. Department of Pharmacology, Faculty of Medicine, University of Malaya, Kuala Lumpur 50603, Malaysia
7. Oral Cancer Research and Coordinating Centre, Faculty of Dentistry, University of Malaya, Kuala Lumpur 50603, Malaysia
Received 2015-11-7; Accepted 2016-3-31; Published 2016-4-26
Abstract
Breast cancer is one of the most common cancers that affect women globally and accounts for ~23% of all cancers diagnosed in women. Breast cancer is also one of the leading causes of death primarily due to late stage diagnoses and a lack of effective treatments. Therefore, discovering protein expression biomarkers is mandatory for early detection and thus, critical for successful therapy. Two-dimensional electrophoresis (2D-E) coupled with lectin-based analysis followed by mass spectrometry were applied to identify potential biomarkers in the secretions of a murine mammary carcinoma cell line. Comparisons of the protein profiles of the murine 4T1 mammary carcinoma cell line and a normal murine MM3MG mammary cell line indicated that cadherin-1 (CDH), collagenase 3 (MMP-13), Viral envelope protein G7e (VEP), Gag protein (GAG) and Hypothetical protein LOC433182 (LOC) were uniquely expressed by the 4T1 cells, and pigment epithelium-derived factor (PEDF) was exclusively secreted by the MM3MG cells. Further analysis by a lectin-based study revealed that aberrant O-glycosylated CDH, N-glycosylated MMP-13 and LOC were present in the 4T1 medium. These differentially expressed N- and O-linked glycoprotein candidates, which were identified by combining lectin-based analysis with 2D-E, could serve as potential diagnostic and prognostic markers for breast cancer.
Keywords: Breast cancer, Murine, Glycosylation, Lectin
Introduction
Breast cancer is a type of molecular disease that is largely caused by mutations of the genes responsible for cell growth and proliferation [1]. Breast cancer can be sporadic or hereditary in nature. The predominant type of breast cancer is sporadic and caused by mutations that occur in somatic cells [2]. The inheritance of germ line mutations, better known as familial or hereditary breast cancer, represents approximately 5-10% of breast cancer cases [3]. Genetic mutations in cancer cells usually involve two major families of genes, i.e., oncogenes and tumor suppressor genes [4, 5].
Breast cancer is a common type of cancer that affects women globally and accounts for 23-29% of all cancers diagnosed in women [6-8]. In 2010, over 207,000 new invasive breast cancer cases were reported in the US, and 39,840 breast cancer patients died [9]. Moreover, 300,000 new cases were reported in 2013, and breast cancer was subsequently classified as the most common cancer in women [8]. According to the National Cancer Registry Report: Malaysia Cancer Statistics - Data and Figures 2007 published by the Ministry of Health Malaysia in 2011 [10], breast cancer was the most common cancer among women in Malaysia in 2007. Breast cancer constitutes 32.1% of the total cancers among women and has an age-standardized incidence rate (ASR) of 29.1 per 100,000 persons. Chinese women have the highest ASR (38.1 per 100,000 population) followed by Indian and Malay women with the ASRs of 33.7 and 25.4 per 100,000 population, respectively. The majority of breast cancer incidences are detected in stage II, and these cases constitute 37% of the total breast cancer diagnoses, followed by stage III (24%), stage I (21%) and stage IV (18%).
Although the mortality rate decreased by 26.58% from 1991 to 2005, breast cancer remains as one of the leading causes of death in women [11]. This high death rate is mainly due to late stage diagnoses and a lack of effective treatments. Therefore, early detection is critical for successful therapy and ultimately the survival of breast cancer patients. Presently, mammograms are one potential detection strategies for breast cancer, but they fail to differentiate benign and malignant lesions [12], and biopsies are required for confirmation [13]. Therefore, different detection strategies have been proposed in the past to evaluate the occurrence of breast cancers. These detections primarily rely on biomarkers and probes [13-17]. Thus, the identification of differentially expressed biomarkers will provide additional strength and hasten early detection. Among the various proposed strategies for searching for biomarkers, proteomic technology appears to be a powerful tool to facilitate the identification of potential disease-associated proteins that can be used for the diagnosis, prognosis, treatment and monitoring of disease [18-21]. Proteomic technology has been used extensively in the study of the pathogeneses of diseases and in the discovery of new biomarkers of disease. However, the biomarkers that are currently used in for the diagnosis and prognosis of breast cancer are not specific or sensitive [22]. There is a need to identify promising tumor markers that can be used individually or in combination with other biomarkers to increase the sensitivities and specificities of the diagnosis and prognostic of breast cancer.
Towards this goal, our study aimed to identify the secreted biomarkers in the culture media of murine mammary carcinoma cell line 4T1. 4T1 cell line is one of only a few breast cancer models with the capacity to metastasize efficiently to sites affected in human breast cancer and the later events that occur at the site of metastasis are most often responsible for patient mortality and morbidity [23]. Study on the secreted protein profile of the cell line could provide new information regarding to the factors involved in directing micro-environmental changes within the tumor that lead to tumor cell dissemination and metastasis, and eventually helps in analysis of cancer progression and evaluation of therapeutics for cancer treatment [24, 25]. Since a biomarker should exhibit relatively high specificity and be expressed at a higher level in the tumor [13], we compared the presence of markers of tumor cells against normal cells.
Furthermore, many of the aberrantly expressed proteins in cancer are acute phase proteins, which are induced upon inflammation [26]. Majority of the acute phase proteins are glycosylated, and it has been shown that alterations in glycosylation may occur in inflammatory and malignant conditions. Therefore, alterations in glycosylation could be potential biomarkers in cancer and have been shown to correlate with disease severity in certain conditions [27]. Special attention was given to the analyses of post-translational modifications using the O- and N-linked glycoprotein profiles of the secreted proteins expressed by the 4T1 cell line.
Materials and Methods
Cell culture
The murine mammary carcinoma cell line 4T1 (ATCC catalogue no.: CRL-2539), and the normal murine mammary cell line MM3MG (ATCC catalogue no.: CRL-6376), were purchased from the American Type Culture Collection (ATCC). The 4T1 and MM3MG cells were maintained in Dulbecco's modified eagle medium (DMEM) growth medium (Invitrogen, California, US) containing 10% fetal bovine serum (Invitrogen, California, US) and cultured at 37oC in a humidified atmosphere containing 5% CO2.
Harvesting of the growth medium
The growth media were removed when the cells were approximately 80% confluent in a 75 cm2 flask. The cells were then washed three times with phosphate buffered saline (PBS) (Invitrogen, California, US) at pH 7.4, and incubated for an additional of 24 h in serum-free DMEM. After 24 h, the media were harvested and centrifuged at 2,000 x g to remove all cell debris and kept at -80oC. Before subjecting the harvested media to two-dimensional electrophoresis (2D-E), the media were concentrated 100-fold using Vivaspin 10,000 molecular weight cut-off concentrators (Sartorius, Goettingen, Germany) and desalted with 2D Clean-Up Kits (GE Healthcare Bio-Sciences, Uppsala, Sweden).
Two-dimensional electrophoresis (2D-E) and silver staining
The 2D-E was performed as previously described [28]. Briefly, 100 µg of concentrated proteins were rehydrated overnight with 250 μl of rehydration buffer (8 M urea, 2 M thiourea, 4% CHAPS, 0.5% pharmalyte, 20 mM dithiothreitol) in 13 cm precast immobilized dry strips at pH 4-7 and pH 3-10 (GE Healthcare Bio-Sciences, Uppsala, Sweden). The strips were then subjected to first-dimensional separation using the Ettan IPGphor II IEF system (GE Healthcare Bio-Sciences, Uppsala, Sweden), followed by the second-dimensional separation at 16°C using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 8-18% gradient gels.
The 2D-E gels were silver stained according to the methods of Heukeshoven and Dernick [29]. For mass spectrometry, the gels were silver stained as described by Shevchenko et al. [30] with modifications.
Image analysis
A GS-710 Imaging Densitometer (Bio-Rad, California, US) and PDQuest® software version 4.7.0 (Bio-Rad, California, US) were used to capture, store and analyze the images of the 2D-E gels. The PDQuest software is able to match identical spots in a series of gels and normalize the gels to compensate for any variations between the gels. The results were normalized to the total density of the gel; the raw quantity of each spot in the gel was divided by the total intensity value over all of the pixels in the image. The normalized spot quantities were expressed as percentages of the volume contributions (vol %) to facilitate the data compilation.
Statistical analysis
All protein concentration values on the gels are presented as the mean of the percentage volume (% volume) ± SE of triplicates of different cell batches. The Student's t-test was used to analyze the significance of the differences between the normal and cancer samples and to examine the correlations between the variables. P-values below 0.05 were considered statistically significant.
Protein identification using mass spectrometry (MS)
Highly resolved spots of interest were excised and subjected to in-gel digestion using a ProteoExtract™ All-in-One Trypsin Digestion Kit (Merck, Darmstadt, Germany). The trypsin-digested peptides were purified and concentrated using pre-packed C-18 Zip Tip Pipette Tips (Millipore, MA, US). The peptides were then mixed with 1 μl of CHCA (5 mg/ml of alpha-cyano-4-hydroxy-cinamic acid in 0.1% trifluoroacetic acid and 50% acetonitrile in deionized distilled water) and spotted onto a matrix-assisted laser desorption/ionization (MALDI) target plate. The mass spectrometric analyses were performed at the Proteomic Centre of the Department of Biological Sciences, National University of Singapore. The peptide mass spectra were obtained using an ABI 4800 Proteomics Analyzer MALDI-TOF/TOF Mass Spectrometer (Applied Biosystems, Framingham, MA, US). The five most intense ions obtained from the MS were subsequently subjected to MS/MS analyses using air with collision energy of 2 kV and a collision gas pressure of ~ 1 x 10-6 Torr. The stop conditions were implemented such that 2000-3000 shots were accumulated based on the quality of the spectra.
For protein identification, the obtained mass spectra were searched against the NCBInr protein database using the MASCOT search engine (version 2.1; Matrix Science, London, UK). The searches were performed with fixed modifications on the carbamidomethylations of the cysteines and variable modifications of the methionine oxidations. The following parameters were used in the MASCOT peptide mass fingerprint search: (i) enzyme: trypsin, (ii) one missed cleavage allowed, (iii) mass value: monoisotopic, (iv) peptide mass tolerance: ± 0.1Da and (v) peptide charge state: 1+. The same parameters were used in the MASCOT ion search, except the peptide mass tolerance and fragment mass tolerance were set to 100 ppm and 0.2 Da, respectively. Search scores above 50 indicated identity or extensive homology (p<0.05).
N-glycosylation analysis using concanavalin A affinity chromatography
Concanavalin A (Con A) is a tetrameric metalloprotein isolated from Canavalia ensiformis (jack bean). Con A coupled to sepharose is routinely used for the separation and purification of N-glycoproteins. To obtain the chromatographic profiles, 10 ml of harvested medium was mixed with 2 ml of Con-A sepharose (GE Healthcare Bio-Sciences, Uppsala, Sweden) and left overnight at 4oC on a rotator. The mixture was then loaded into a column (0.8 cm X 4 cm; BioRad Laboratories, Hercules, CA, US), equilibrated and washed with equilibration buffer (20 mM Tris-HCl, 0.5 M NaCl, pH 7.4) to remove the non-glycosylated and O-glycosylated proteins. The bound N-glycoproteins were then eluted with 0.3 M of methyl-α-D-glucopyranoside. The chromatographic process was monitored at an absorbance of 280 nm. The eluted fractions were concentrated 100 times using Vivaspin 10,000 molecular weight cut-off concentrators. The concentrated fractions were further desalted and subjected to 2D-E analyses.
Western blotting and detection of the O-linked glycosylation using champedak galactose binding (CGB) lectin
The 2D-E gels from the growth media were transferred onto a nitrocellulose membrane (0.45 μm) using a Multiphor II NovaBlot Kit (GE Healthcare Bio-Sciences, Uppsala, Sweden). The blotted membrane was blocked with 5% skim milk in Tween TRIS-buffered saline (TTBS) for 1 h at room temperature, washed 3 times with the same buffer and then incubated with CGB lectin conjugated to horseradish peroxidase (HRP) at a concentration of approximately 1 μg/ml at 4oC overnight. The purity and specificity of the interaction of CGB lectin with the O-glycosylated proteins have been described previously [31]. After the incubation, the membrane was washed twice and developed using freshly prepared substrate solution consisting of 0.03 g of 3,3'-diaminobenzidine (Dako, Glostrup, Denmark) and 50 ul of H2O2 in 50 ml of Tris-buffered saline. The membrane was washed twice with deionized distilled water to terminate the reaction. The developed membrane was then air-dried and scanned using a GS-710 Imaging densitometer (Bio-Rad, California, US).
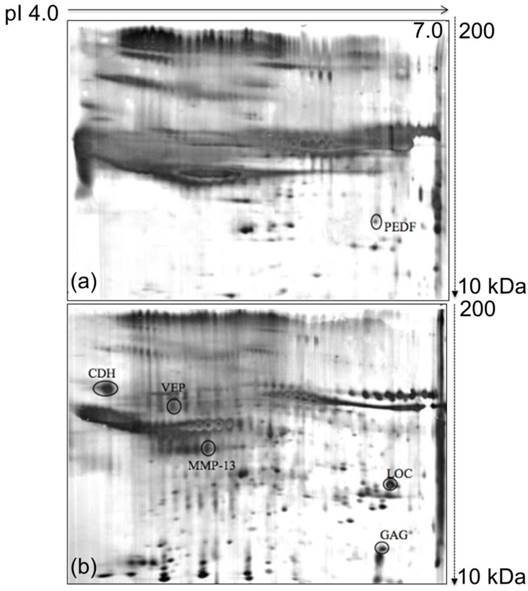
Comparison of 2D-E protein profiles derived from the growth media of MM3MG and 4T1 cells. (a) Typical 2D-E growth medium profile of the MM3MG cell line. (b) Typical 2D-E growth medium profile of the 4T1 cell line. The growth media of the 4T1 and MM3MG cells were subjected to 2D-E and silver stained. PEDF was detected in the MM3MG growth medium, and CDH, MMP-13, VEP, GAG and LOC were clearly resolved in the 4T1 growth medium profile.
Results and Discussion
The focus of this study was the analysis of the proteins involved in breast cancer progression because breast cancer is one of the major cancers that is reported all over the world as difficult to detect in the earlier stages. To fill this lacuna, the proteins secreted by the murine 4T1 mammary carcinoma cells were analyzed and compared with those of the normal murine MM3MG cells. The 4T1 cell line was derived from BALB/c mice, and the cancer arose spontaneously. The MM3MG cell line is a normal BALB/c mammary epithelial cell line. Separation of the 4T1 and MM3MG media by 2D-E produced clear and resolved protein profiles that consisted of diverse secreted proteins. Furthermore, differentially expressed proteins were identified in the comparisons of the observed protein biomarkers from the 4T1 cells against those of the MM3MG cells (Figure 1a & b).
Imaging analyses on the 2D-E resolved spot profiles
The 2D-E gels of the growth media of the 4T1 and MM3MG cells were scanned using a GS-710 imaging densitometer and analyzed using PDQuest™ 2D gel analysis software. Comparative analysis revealed 6 spots of differentially regulated proteins in the media of the 4T1 and MM3MG cells. Five spots were preferentially expressed in the 4T1 cells, and one was preferentially expressed in the MM3MG cells. These spots were further subjected to mass spectrometric analysis for identification.
Identification of secreted protein from murine 4T1 mammary carcinoma cell line
The identification of the protein spots was performed using an ABI 4800 proteomics analyzer MALDI-TOF/TOF mass spectrometer. The obtained mass spectra were searched against the NCBInr protein database using the MASCOT search engine. The parameters was set as follows: one missed cleavage allowed in the trypsin digestion; limited to Mus musculus; monoisotopic mass value; ±0.1 Da peptide mass tolerance; and 1+ peptide charge state. The mass spectra of the digested samples are shown in Figure S1. Search scores greater than 50, which indicated extensive homology, were obtained for the tested protein spots. The protein specifically secreted by the MM3MG cell line was identified by MS/MS as Pigment epithelium-derived factor precursor (PEDF) (Figure 1a). Additionally, the proteins detected in 4T1 medium consisted of Cadherin-1 (CDH), Collagenase 3 Preproprotein (MMP-13), Viral envelope protein G7e (VEP), Gag protein (GAG) and Hypothetical protein LOC433182 (LOC) (Table 1; Figure 1b).
Images of the glycoprotein profiles derived from the growth media of the 4T1 cells. (a) Typical 2D-E N-glycoprotein profile of the 4T1 growth medium. (b) Typical representative O-glycoprotein profile of the 4T1 growth medium. The growth media harvested from the 4T1 cells were subjected to Con A and HRP-conjugated CGB lectin-based analyses. LOC and MMP-13 were detected as N-glycoproteins, and CDH was detected as an O-glycoprotein in the 4T1 growth medium.

The correlations of the aberrantly expressed proteins identified in this in vitro study with breast cancer were further investigated with glycosylation analysis. Glycosylation is a type of extensive post-translational modification that can significantly alter protein function and thus modify cellular characteristics. Several pieces of evidence have indicated that abnormal glycosylation can contribute to the development of cancer [32, 33]. Therefore, aberrantly glycosylated proteins might be useful for diagnostic purposes. In this study, post-translational modification analyses were performed for both N- and O- linked glycoproteins in the growth media of the 4T1 and MM3MG cell lines (Figure 2a, 2b and 4). The post-translational protein profiles were generated by detection with Con A and HRP-conjugated CGB lectins.
Mass spectrometric identification of the protein spots from the 4T1 and MM3MG growth media using the MASCOT search engine and the NCBI database.
| Spot Name | Protein Name | Mascot Accession number | Theoretical pI | Theoretical Mass (Da) | No. of Peaks Matched | Search Score |
|---|---|---|---|---|---|---|
| CDH | Cadherin-1 | gi|6753374 | 4.70 | 98736 | 4 | 197 |
| MMP-13 | Collagenase 3 preproprotein | gi|6678896 | 5.12 | 54376 | 2 | 82 |
| VEP | Viral envelope Protein G7e | gi|16118495 | 8.39 | 67039 | 2 | 87 |
| GAG | Gag protein | gi|34391479 | 7.63 | 60596 | 3 | 267 |
| LOC | Hypothetical Protein LOC433182 | gi|70794816 | 6.37 | 47453 | 7 | 557 |
| PEDF | Pigment epithelium-derived factor | gi|117606335 | 6.48 | 46262 | 3 | 184 |
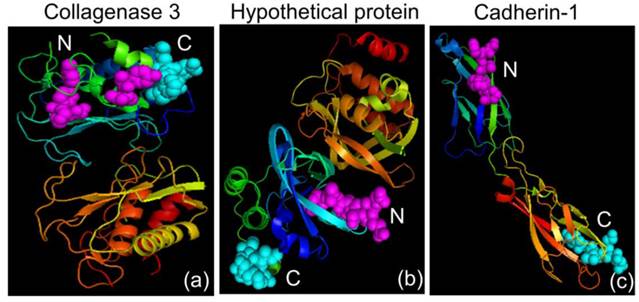
Three-dimensional crystal structures. (a) Collagenase 3 Preproprotein (PDB accession code: 1CXV), (b) Hypothetical Protein (PDB accession code: 215T) and (c) Cadherin-1 (PDB accession code: 2QVF). The N and C terminals are shown in magenta and cyan, respectively.
N-glycosylated proteins from the murine 4T1 mammary carcinoma cell line
When the 4T1 and MM3MG media were subjected to Con A chromatography and subsequently analyzed by 2D-E, distinctly different profiles were produced. Strikingly, no distinct N-glycoproteins were detected in the MM3MG medium, whereas the 4T1 cells expressed N-glycosylated LOC and MMP-13 (Figure 2a).
LOC is a protein sequence that has been obtained from analyses of mouse cDNA sequences [34]. Little information is available regarding the protein's functionality, but three-dimensional structural information about the different hypothetical LOC proteins is available (Example, PDB accession code: 215T; Figure 3b). The detection of LOC in the medium of the 4T1 cells and not in the MM3MG medium might suggest that this protein sequence has a role in tumorigenesis.
Collagenase 3 is also known as matrix metalloproteinase-13 (MMP-13) and is member of the matrix metalloproteinase family, which breaks down the proteins of the extracellular matrix [35]. Matrix metalloproteinase plays important roles in tissue remodeling, tissue repair, embryogenesis and cell differentiation [36, 37]. The matrix metalloproteinase family can be classified into subgroups of collagenases, gelatinases, novel matrix metalloproteinases, membrane-type matrix metalloproteinases and stromelysins according to substrate specificity and structure [36, 37]. MMP-13 is able to degrade types I, II, III, IV, IX, X and XIV collagens, tenascin, fibronectin and aggrecan core protein [38-41]. The wide range of the substrate specificity of MMP-13 makes it an effective tool for tumor cells invasion. The expression of MMP-13 in relation to cancer has been detected in breast cancer [42], squamous cell carcinomas of the head and neck [43], chondrosarcomas [44] and colorectal cancer [45]. Structural information about mouse collagenase is easily accessible (PDB accession code: 1CXV; Figure 3a) and provides details about the specificity of collagenase and drug design [46]. In the present study, we detected an abnormal secretion of MMP-13 in the 4T1 medium and not in the MM3MG medium. The secretion of MMP-13 from the 4T1 cells may play a role in the breakdown of the extracellular matrix surrounding the cells and thus aid in tumor invasion and progression [47]. We also observed the presence of MMP-13 as an N-glycoprotein in the 4T1 medium. A previous study by Knauper et al. [40] reported the presence of an N-glycosylation site on the human MMP-13. However, no study has demonstrated the presence of an N-glycosylation site in the mouse MMP-13.
O-glycosylated proteins from the murine 4T1 mammary carcinoma cell line
Examinations of the secreted O-glycosylated proteins in the 4T1 and MM3MG media were performed using blotting with HRP-conjugated CGB lectin. Notably, Cadherin-1 (CDH) was detected as an O-glycoprotein in the 4T1 media (Figure 2b).
CDH is also known as L-CAM epithelial cadherin (E-cadherin) and uvomorulin and has been reported to be a calcium-dependent glycoprotein that regulates cell-cell adhesion [48]. CDH is usually expressed on the surfaces of epithelial cells in adherens junctions [49]. CDH is associated with gland formation and the stratification and polarization of epithelial cells [50]. CDH is encoded by the CDH1 gene located on chromosome 16q22.1, which is known to be a tumor suppressor gene [51, 52]. The loss of CDH protein has been associated with increases in the invasiveness and metastasis of tumors [52, 53]. Diverging levels of CDH expression have been associated with cancer invasiveness. Decreased expression of CDH has been reported in tumors of the ovary, esophagus and stomach [54-56]. In contrast, elevated CDH levels have been observed in inflammatory breast cancer and epithelial ovarian cancers [57, 58]. The structural information about the mouse CDH is currently available in the RCSB Protein Data Bank (PDB accession code: 2QVF; Figure 3c). In this study, we detected CDH in the secretome of the 4T1 cell line but not in that of the MM3MG cell line. We also detected an aberrant glycosylation of CDH in the 4T1 secretome. CDH was detected as an O-glycoprotein in the medium of the 4T1 cells. O-glycosylated CDH has previously been reported in the MDA-MB-231 breast cancer cell line [59]. Another study also reported that induced O-glycosylated CDH in a breast cancer cell line is linked to the blocking of cell surface transport and reduced intercellular adhesion [60]. We postulate that the aberrantly glycosylated CDH is associated with the increased invasiveness and metastatic ability of the cancer cell line via reductions in the intercellular adhesion force.
Suppression of pigment epithelium-derived factor in the murine 4T1 mammary carcinoma cell line
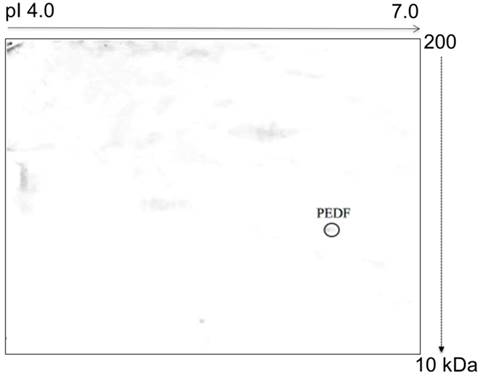
Glycosylation analysis of the 4T1 cells demonstrated 3 different post-translationally modified N- and O-linked MMP-13, LOC and CDH glycoproteins. These glycoproteins were not observed in the normal MM3MG cells. In contrast, O-glycosylation analysis of the media from the MM3MG cells revealed Pigment epithelium-derived factor (PEDF) (Figure 4).
PEDF is a 50-kDa secreted glycoprotein that belongs to the serine protease inhibitor superfamily [61, 62]. PEDF was initially discovered in the secretions of retinal pigment epithelial cells [63] but was later found to be expressed by several tissues, including the plasma, bone, prostate, heart, lungs, brain and spinal cord [64]. PEDF is a potent inhibitor of angiogenesis that neutralizes the pro-angiogenic effect of vascular endothelial growth factor [65, 66]. Moreover, PEDF also acts as an anti-angiogenic agent, an anti-inflammatory agent [67], an anti-fibrosis agent [68] and an anti-vasopermeability agent [69]. Thus, PEDF is a multifunctional protein.
In the current study, we detected the presence of PEDF in the medium of the MM3MG cell line but not in that of the 4T1 cells. PEDF has been shown to possess anti-tumor properties mediated by the induction of apoptosis, the promotion of tumor differentiation and the inhibition of angiogenesis [65, 70-72]. Other studies have also reported significant reductions in PEDF expression in breast cancer [73], ovarian cancer [74], prostate cancer [75] and pancreatic adenocarcinoma [76]. Hence, the lack of PEDF in the 4T1 medium suggested that the tumor cells were able to suppress PEDF expression to aid tumor progression. Conversely, we detected the PEDF as an O-glycoprotein in the MM3MG medium. The finding of O-glycosylated PEDF is consistent with previous studies that have revealed an O-glycosylation site in the human PEDF [77].
Non-glycosylated proteins from the murine 4T1 mammary carcinoma cell line
In addition to the N- and O-glycosylated proteins, there were two non-glycosylated proteins that were detected in the 4T1 cell, i.e., viral envelope protein G7e (VEP) and Gag protein (GAG).
VEP is a protein that is encoded by novel gene G7e. This gene is transcribed in lymphoid tissues to produce a 3 kb mRNA. The G7e gene is located in a region in which recombination preferentially occurs, and several disease susceptibility genes have been mapped to the same region [78]. Studies of this protein since its discovery are limited. The detection of VEP protein expression in the medium of the 4T1 cells is suggestive of this protein's role in tumorigenesis. Because the gene that encodes the VEP protein is located in close proximity to a recombinant region, the integration of this gene into a cancer causing gene may form a new recombinant protein that could play a certain role in tumorigenesis.
GAG is a protein that is found in mouse mammary tumor virus and coordinates the assembly of completed viral capsids in the cytoplasm that are subsequently transported out of the cells [79]. In the current study, we detected the presence of the GAG protein in the medium of the 4T1 cell line. A previous study reported an elevation in gene expression related to the envelope of the murine acquired immunodeficiency syndrome (MAIDS) virus following burn injury and the inflammatory response [80]. Our data revealed findings that were similar to those of a previous study that found that the expression of viral-related GAG is detectable in the secretions of the 4T1 cell line. This finding suggests that the reactivation of defective or dormant virus-related sequences might be affected by pathophysiological states such as cancer. The expression of this virus-related sequence might affect the transcriptional activities of adjacent genes in the integration region, which may then produce new recombinant proteins and induce the progression of cancer.
Typical representative image of an O-glycoprotein profile derived from the MM3MG growth medium. The harvested growth media were subjected to 2D-E, blotted and developed with HRP-conjugated CGB lectin. PEDF was detected as an O-glycoprotein in the MM3MG growth medium.
Conclusions
Breast cancer is one of the leading causes of death primarily due to late stage diagnoses and the lack of effective treatments. To enhance the survival rate, this study identified differentially expressed and glycosylated proteins in the secretion of a murine mammary cancer cell line. We employed a proteomics approach using 2D-E coupled with lectin-based analysis to identify the aberrantly expressed N- and O-linked glycoproteins in the secretome. Subsequent analyses of the murine 4T1 tumor cells revealed the unique expressions of the CDH, MMP-13, VEP, GAG and LOC proteins in the medium of the 4T1 cell line and not in the medium of the MM3MG cells. However, the unique expression of PEDF in the medium of the MM3MG cells was noted. The lectin-based analysis revealed that CDH was aberrantly O-glycosylated and that MMP-13 and LOC were differentially N-glycosylated in the 4T1 growth medium. The findings of the current study identified interesting potential biomarkers for the diagnosis and prognosis of breast cancer. Therefore, the use of lectin-based analysis coupled with 2D-E provided a more sensitive method for detecting and verifying differentially glycosylated proteins in breast cancer samples. Further studies have to be done on the identified biomarkers to reveal the actual role/functions of these biomarkers in breast cancer.
Supplementary Material
Figure S1.
Acknowledgements
This study was supported by University of Malaya Research Grants (UMRG) RG454-12HTM and University of Malaya (UM) High Impact Research (HIR) MoE Grants UM.C/625/1/HIR/MOE/DENT/09 and UM.C/625/1/HIR/MOHE/MED/16/5 from the Ministry of Education Malaysia.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ingvarsson S. Genetics of breast cancer. Drugs Today (Barc). 2004;40:991-1002
2. Kenemans P, Verstraeten R, Verheijen R. Oncogenic pathways in hereditary and sporadic breast cancer. Maturitas. 2004;49:34-43
3. Pavelić K, Gall-Trošelj K. Recent advances in molecular genetics of breast cancer. J Mol Med (Berl). 2001;79:566-73
4. Macdonald F, Ford C, Casson A. Molecular Biology of Cancer. United Kingdom: Taylor & Francis. 2004
5. Schulz WA. Molecular biology of human cancers: an advanced student's textbook. Dordrecht: Springer Science & Business Media. 2005
6. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-917
7. Sasco AJ, Kaaks R, Little RE. Breast cancer: occurrence, risk factors and hormone metabolism. Expert Rev Anticancer Ther. 2003;3:546-62
8. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29
9. American Cancer Society. Breast Cancer Facts and Figures 2009-2010. Atlanta: American Cancer Society, Inc. 2009
10. Zainal Ariffin O, Nor Saleha IT. National Cancer Registry Report 2007. Malaysia: Ministry of Health. 2011
11. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225-49
12. Kopans D. Breast Imaging. 3 ed. Philadelphia: Lippincott Williams & Wilkins. 2007
13. Hathaway H, Butler K, Adolphi N, Lovato D, Belfon R, Fegan D. et al. Detection of breast cancer cells using targeted magnetic nanoparticles and ultra-sensitive magnetic field sensors. Breast Cancer Res. 2011;13:R108
14. Musayev J, Altiner C, Adiguzel Y, Kulah H, Eminoglu S, Akin T. Capturing and detection of MCF-7 breast cancer cells with a CMOS image sensor. Sens Actuators A Phys. 2014;215:105-14
15. Nguyen CV, Saraf RF. Tactile Imaging of an Imbedded Palpable Structure for Breast Cancer Screening. ACS Appl Mater Interfaces. 2014;6:16368-74
16. Tehrani Z, Burwell G, Azmi MM, Castaing A, Rickman R, Almarashi J. et al. Generic epitaxial graphene biosensors for ultrasensitive detection of cancer risk biomarker. 2D Mater. 2014:1 025004
17. Tan A-A, Phang W-M, Gopinath SC, Hashim OH, Kiew LV, Chen Y. Revealing Glycoproteins in the Secretome of MCF-7 Human Breast Cancer Cells. Biomed Res Int. 2015. 2015
18. Hunt DF. Personal commentary on proteomics. J Proteome Res. 2002;1:15-9
19. Chen Y, Azman SN, Kerishnan JP, Zain RB, Chen YN, Wong Y-L. et al. Identification of Host-Immune Response Protein Candidates in the Sera of Human Oral Squamous Cell Carcinoma Patients. PLoS One. 2014;9:e109012
20. Gopinath SCB, Wadhwa R, Kumar PKR. Expression of Noncoding Vault RNA in Human Malignant Cells and Its Importance in Mitoxantrone Resistance. Mol Cancer Res. 2010;8:1536-46
21. Gopinath SCB, Matsugami A, Katahira M, Kumar PKR. Human vault-associated non-coding RNAs bind to mitoxantrone, a chemotherapeutic compound. Nucl Acids Res. 2005;33:4874-81
22. Arslan N, Serdar M, Deveci S, Ozturk B, Narin Y, Ilgan S. et al. Use of CA15-3, CEA and prolactin for the primary diagnosis of breast cancer and correlation with the prognostic factors at the time of initial diagnosis. Ann Nucl Med. 2000;14:395-9
23. Tao K, Fang M, Alroy J, Sahagian GG. Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer. 2008;8:228
24. Munzone E, Colleoni M. Clinical overview of metronomic chemotherapy in breast cancer. Nat Rev Clin Oncol. 2015;12:631-44
25. Zardavas D, Irrthum A, Swanton C, Piccart M. Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol. 2015;12:381-94
26. Gruys E, Toussaint M, Niewold T, Koopmans S. Review: Acute phase reaction and acute phase proteins. J Zhejiang Univ Sci B. 2005;6:1045
27. McCarthy C, Saldova R, Wormald MR, Rudd PM, McElvaney NG, Reeves EP. The role and importance of glycosylation of acute phase proteins with focus on alpha-1 antitrypsin in acute and chronic inflammatory conditions. J Proteome Res. 2014;13:3131-43
28. Chen Y, Lim B-K, Peh S-C, Abdul-Rahman PS, Hashim OH. Profiling of serum and tissue high abundance acute-phase proteins of patients with epithelial and germ line ovarian carcinoma. Proteome Sci. 2008;6:20
29. Heukeshoven J, Dernick R. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis. 1985;6:103-12
30. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850-8
31. Rahman MA, Karsani SA, Othman I, Rahman PSA, Hashim OH. Galactose-binding lectin from the seeds of champedak (Artocarpus integer): sequences of its subunits and interactions with human serum O-glycosylated glycoproteins. Biochem Biophys Res Commun. 2002;295:1007-13
32. Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525-46
33. Tuccillo FM, de Laurentiis A, Palmieri C, Fiume G, Bonelli P, Borrelli A. et al. Aberrant glycosylation as biomarker for cancer: focus on CD43. Biomed Res Int. 2014. 2014
34. Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS. et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899-903
35. Frederick Woessner J. The Family of Matrix Metalloproteinasesa. Ann N Y Acad Sci. 1994;732:11-21
36. Birkedal-Hansen H, Moore W, Bodden M, Windsor L, Birkedal-Hansen B, DeCarlo A. et al. Matrix metalloproteinases: a review. Crit Rev Oral Biol Med. 1993;4:197-250
37. Kähäri VM, Saarialho-Kere U. Matrix metalloproteinases in skin. Exp Dermatol. 1997;6:199-213
38. Fosang AJ, Last K, Knäuper V, Murphy G, Neame PJ. Degradation of cartilage aggrecan by collagenase-3 (MMP-13). FEBS Lett. 1996;380:17-20
39. Knäuper V, Cowell S, Smith B, López-Otin C, O'Shea M, Morris H. et al. The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J Biol Chem. 1997;272:7608-16
40. Knäuper V, López-Otin C, Smith B, Knight G, Murphy G. Biochemical characterization of human collagenase-3. J Biol Chem. 1996;271:1544-50
41. Mitchell PG, Magna HA, Reeves LM, Lopresti-Morrow LL, Yocum SA, Rosner PJ. et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J Clin Invest. 1996;97:761-8
42. Chang H-J, Yang M-J, Yang Y-H, Hou M-F, Hsueh E-J, Lin S-R. MMP13 is potentially a new tumor marker for breast cancer diagnosis. Oncol Rep. 2009;22:1119-27
43. Johansson N, Airola K, Grénman R, Kariniemi A-L, Saarialho-Kere U, Kähäri V. Expression of collagenase-3 (matrix metalloproteinase-13) in squamous cell carcinomas of the head and neck. Am J Pathol. 1997;151:499-508
44. Uría JA, Balbín M, López JM, Alvarez J, Vizoso F, Takigawa M. et al. Collagenase-3 (MMP-13) expression in chondrosarcoma cells and its regulation by basic fibroblast growth factor. Am J Pathol. 1998;153:91-101
45. Huang M-Y, Chang H-J, Chung F-Y, Yang M-J, Yang Y-H, Wang J-Y. et al. MMP13 is a potential prognostic marker for colorectal cancer. Oncol Rep. 2010;24:1241-7
46. Botos I, Meyer E, Swanson SM, Lemaı̂tre V, Eeckhout Y, Meyer EF. Structure of recombinant mouse collagenase-3 (MMP-13). J Mol Biol. 1999;292:837-44
47. McLeod H, Murray G. Tumour markers of prognosis in colorectal cancer. Br J Cancer. 1999;79:191-203
48. Gallin WJ, Sorkin BC, Edelman GM, Cunningham BA. Sequence analysis of a cDNA clone encoding the liver cell adhesion molecule, L-CAM. Proc Natl Acad Sci U S A. 1987;84:2808-12
49. Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345-57
50. Bracke M, Van Roy F, Mareel M. The E-cadherin/catenin complex in invasion and metastasis. Attempts to Understand Metastasis Formation I. Springer. 1996:123-61
51. Berx G, Cleton-Jansen A, Nollet F, De Leeuw W, Van de Vijver M, Cornelisse C. et al. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995;14:6107-15
52. Vleminckx K, Vakaet L, Mareel M, Fiers W, Van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107-19
53. Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A. et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173-85
54. Handschuh G, Candidus S, Luber B, Reich U, Schott C, Oswald S. et al. Tumour-associated E-cadherin mutations alter cellular morphology, decrease cellular adhesion and increase cellular motility. Oncogene. 1999;18:4301-12
55. Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806-11
56. Tang AM, Amagai M, Granger LG, Stanley JR, Udey MC. Adhesion of Epidermal Langerhans Cells to Keratinocytes Mediated by E-Cadherin. Nature. 1993;361:82-5
57. Kleer CG, van Golen KL, Braun T, Merajver SD. Persistent E-cadherin expression in inflammatory breast cancer. Mod Pathol. 2001;14:458-64
58. Sundfeldt K. Cell-cell adhesion in the normal ovary and ovarian tumors of epithelial origin; an exception to the rule. Mol Cell Endocrinol. 2003;202:89-96
59. Vester-Christensen MB, Halim A, Joshi HJ, Steentoft C, Bennett EP, Levery SB. et al. Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc Natl Acad Sci U S A. 2013;110:21018-23
60. Zhu W, Leber B, Andrews DW. Cytoplasmic O-glycosylation prevents cell surface transport of E-cadherin during apoptosis. EMBO J. 2001;20:5999-6007
61. Becerra SP. Structure-function studies on PEDF. A noninhibitory serpin with neurotrophic activity. Adv Exp Med Biol. 1997;425:223-37
62. Becerra SP, Sagasti A, Spinella P, Notario V. Pigment Epithelium-derived Factor Behaves Like a Noninhibitory Serpin. Neurotrophic activity does not require the serpin reactive loop. J Biol Chem. 1995;270:25992-9
63. Barnstable CJ, Tombran-Tink J. Neuroprotective and antiangiogenic actions of PEDF in the eye: molecular targets and therapeutic potential. Prog Retin Eye Res. 2004;23:561-77
64. Filleur S, Nelius T, De Riese W, Kennedy R. Characterization of PEDF: a multi-functional serpin family protein. J Cell Biochem. 2009;106:769-75
65. Dawson D, Volpert O, Gillis P, Crawford S, Xu H-J, Benedict W. et al. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245-8
66. King GL, Suzuma K. Pigment-epithelium-derived factor-a key coordinator of retinal neuronal and vascular functions. N Engl J Med. 2000;342:349-51
67. Zhang SX, Wang JJ, Gao G, Shao C, Mott R, Ma J-x. Pigment epithelium-derived factor (PEDF) is an endogenous antiinflammatory factor. FASEB J. 2006;20:323-5
68. Matsuoka M, Ogata N, Otsuji T, Nishimura T, Takahashi K, Matsumura M. Expression of pigment epithelium derived factor and vascular endothelial growth factor in choroidal neovascular membranes and polypoidal choroidal vasculopathy. Br J Ophthalmol. 2004;88:809-15
69. Amaral J, Fariss RN, Campos MM, Robison WG, Kim H, Lutz R. et al. Transscleral-RPE permeability of PEDF and ovalbumin proteins: implications for subconjunctival protein delivery. Invest Ophthalmol Vis Sci. 2005;46:4383-92
70. Ek ET, Dass CR, Choong PF. PEDF: a potential molecular therapeutic target with multiple anti-cancer activities. Trends Mol Med. 2006;12:497-502
71. Ek ET, Dass CR, Choong PF. Pigment epithelium-derived factor: a multimodal tumor inhibitor. Mol Cancer Ther. 2006;5:1641-6
72. Fernández-García NI, Volpert OV, Jimenez B. Pigment epithelium-derived factor as a multifunctional antitumor factor. J Mol Med (Berl). 2007;85:15-22
73. Cai J, Parr C, Watkins G, Jiang WG, Boulton M. Decreased pigment epithelium-derived factor expression in human breast cancer progression. Clin Cancer Res. 2006;12:3510-7
74. Cheung LW, Au SC, Cheung AN, Ngan HY, Tombran-Tink J, Auersperg N. et al. Pigment epithelium-derived factor is estrogen sensitive and inhibits the growth of human ovarian cancer and ovarian surface epithelial cells. Endocrinology. 2006;147:4179-91
75. Halin S, Wikström P, Rudolfsson SH, Stattin P, Doll JA, Crawford SE. et al. Decreased pigment epithelium-derived factor is associated with metastatic phenotype in human and rat prostate tumors. Cancer Res. 2004;64:5664-71
76. Uehara H, Miyamoto M, Kato K, Ebihara Y, Kaneko H, Hashimoto H. et al. Expression of pigment epithelium-derived factor decreases liver metastasis and correlates with favorable prognosis for patients with ductal pancreatic adenocarcinoma. Cancer Res. 2004;64:3533-7
77. Simonovic M, Gettins PG, Volz K. Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor. Proc Natl Acad Sci U S A. 2001;98:11131-5
78. Snoek M, van Dinten L, van Vugt H. A novel gene, G7e, resembling a viral envelope gene, is located at the recombinational hot spot in the class III region of the mouse MHC. Genomics. 1996;38:5-12
79. Beyer AR, Bann DV, Rice B, Pultz IS, Kane M, Goff SP. et al. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein L9. J Virol. 2013;87:1069-82
80. Cho K, Adamson LK, Greenhalgh DG. Induction of murine AIDS virus-related sequences after burn injury. J Surg Res. 2002;104:53-62
Author contact
![]() Corresponding author: E-Mail address: chenyengedu.my; Phone number: (+60) 3-79676470
Corresponding author: E-Mail address: chenyengedu.my; Phone number: (+60) 3-79676470

 Global reach, higher impact
Global reach, higher impact