3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(12):937-945. doi:10.7150/ijms.11605 This issue Cite
Research Paper
Collagen Triple Helix Repeat Containing-1 (CTHRC1) Expression in Oral Squamous Cell Carcinoma (OSCC): Prognostic Value and Clinico-Pathological Implications
1. Oral Cancer Research and Coordinating Centre (OCRCC), Faculty of Dentistry, University of Malaya, Kuala Lumpur, Malaysia;
2. Department of Oro-Maxillofacial Surgical and Medical Sciences, Faculty of Dentistry, University of Malaya, Kuala Lumpur, Malaysia;
3. Department of Oral and Maxillofacial Surgery, Hospital Tengku Ampuan Rahimah, Selangor Darul Ehsan, Klang, Malaysia;
4. Department of Oral Surgery, Hospital Umum Kuching, Sarawak, Malaysia;
5. Department of Oral and Maxillofacial Surgery, Hospital Kuala Lumpur, Kuala Lumpur, Malaysia;
6. Oral Cancer Research Team, Cancer Research Initiatives Foundation (CARIF), Selangor Darul Ehsan, Malaysia.
Received 2015-1-16; Accepted 2015-8-17; Published 2015-11-1
Abstract
BACKGROUND: Collagen Triple Helix Repeat Containing 1 (CTHRC1) is a protein often found to be over-expressed in various types of human cancers. However, correlation between CTHRC1 expression level with clinico-pathological characteristics and prognosis in oral cancer remains unclear. Therefore, this study aimed to determine mRNA and protein expression of CTHRC1 in oral squamous cell carcinoma (OSCC) and to evaluate the clinical and prognostic impact of CTHRC1 in OSCC.
METHODS: In this study, mRNA and protein expression of CTHRC1 in OSCCs were determined by quantitative PCR and immunohistochemistry, respectively. The association between CTHRC1 and clinico-pathological parameters were evaluated by univariate and multivariate binary logistic regression analyses. Correlation between CTHRC1 protein expressions with survival were analysed using Kaplan-Meier and Cox regression models.
RESULTS: Current study demonstrated CTHRC1 was significantly overexpressed at the mRNA level in OSCC. Univariate analyses indicated a high-expression of CTHRC1 that was significantly associated with advanced stage pTNM staging, tumour size ≥ 4 cm and positive lymph node metastasis (LNM). However, only positive LNM remained significant after adjusting with other confounder factors in multivariate logistic regression analyses. Kaplan-Meier survival analyses and Cox model demonstrated that patients with high-expression of CTHRC1 protein were associated with poor prognosis and is an independent prognostic factor in OSCC.
CONCLUSION: This study indicated that over-expression of CTHRC1 potentially as an independent predictor for positive LNM and poor prognosis in OSCC.
Keywords: Oral cancer, OSCC, CTHRC1, Lymph node metastasis, poor prognosis
Introduction
Oral cancer is a global health problem that causes fear, morbidity and mortality and poses major challenge in many parts of the world. In 2008, approximately 263,900 new oral cancer cases were reported and approximately 128,000 deaths were recorded globally [1]. Oral cavity is one of the most common site of cancer and accounts for about one-third of all cancer in Southern Asian countries such as India, Bangladesh, Pakistan and Sri Lanka [1, 2]. Within the Malaysian population, oral cancer has been reported by National Cancer Registry (NCR) as the 22nd and 15th most common type of cancer in males and females respectively in year 2003-2005 [3].
Generally, most cases of oral cancers are of oral squamous cell carcinoma (OSCC) subtype [4]. As with other subtypes of oral cancer, OSCC also frequently leads to dysfunction of mastication, deglutition and speech, resulting in poor quality of life. Current treatment for OSCCs involves a combination of surgical therapy, chemotherapy, and radiotherapy, all of which have different disadvantages such as poor respond to treatment, and disease can recur or metastasize during the treatment period [5-7]. Despite the clinical advances in the management of cancers, survival for oral cancer specifically has not improved significantly over the past several decades with high mortality rate and five-year survival of only ~50% [8]. In recent years, research in the field of cancer biology has advanced where candidate genes had been identified as diagnostic and prognostic biomarkers for several cancers such as KIT and cytokeratins for gastrointestinal and breast cancer respectively [9].
Collagen triple helix repeat containing-1 (CTHRC1) is a 30kDa secreted and promigratory protein [10] which is highly conserved from lower chordates to mammals and was first found to be expressed during rat tissue repair process [11, 12]. Studies had shown that CTHRC1 protein expression levels were increased in injured and diseased arteries, suggesting that this gene contributes to tissue repair in vascular remodelling in response to injury, and thus promotes cell migration ability. It was also proposed that CTHRC1 could induce abnormal angiogenesis which is an integral hallmark of cancer [13, 14]. Angiogenesis is a critical response to tissue injury [15] and it is activated transiently. In contrast, angiogenesis is almost always kept activated during tumour progression for the survival of the tumour.
Through an array CGH study previously, a copy number gain in chromosome 8q11.1-q24.4 [16] was identified in 54% of OSCC. One of the genes located at this region was CTHRC1. This similar result was in concordance with a recent study which reported the copy number gain of 8q in 53% of the hepatocellular tissues and identified CTHRC1 as a novel HCC-related gene [17]. In addition, elevated expression of CTHRC1 has been observed in various types of human solid cancers such as lung, thyroid, breast, ovarian, cervix, liver, and pancreas [12]. A recent study revealed that over-expression of CTHRC1 in pancreatic cancer tissues plays a critical role in cell migration, adhesiveness and tumourigenesis [18]. Furthermore, aberrant expression of CTHRC1 in gastric carcinogenesis was also reported to promote tumour cell invasion and metastasis [19]. Recently, CTHRC1 has been reported to be over-expressed in OSCC due to hyper-glycosylation and transcriptional activation by canonical Wnt pathway. Over-expression of CTHRC1 in OSCC cell lines stabilized the Wnt-Fzd complex which activates the non-canonical Wnt/PCP signaling pathway [20], which enhanced OSCC migration [21].
As the over-expression of CTHRC1 in cancer cells have been linked to cell invasion and migration [18, 21, 22], it is likely that CTHRC1 could play an important role in driving metastasis. Cervical lymph node metastasis is one of the important clinico-pathological parameters in determining the outcome of OSCC patients [23] and this status is highly correlated with patient survival [24]. The accuracy of the prediction of cervical lymph node metastasis status in OSCC would help clinicians to predict the prognostic outcome of the patients and thus plan the appropriate treatment for oral cancer patients. However, the expression of CTHRC1 in oral cavity cancers and its correlation to clinico-pathological parameters and prognosis remains unclear. The current study aims to determine the expression levels of CTHRC1 in OSCC, and to evaluate its clinical and prognostic significance, which could lead to an improvement in prognosis prediction and development of treatment strategies for OSCC patients.
Materials and Methods
Specimen and data collection
For determination of mRNA expression level of CTHRC1, cDNA samples from 45 OSCC and 5 normal oral mucosal tissues were included in this study. All tissues were earlier snap-frozen and stored in liquid nitrogen at the Malaysian Oral Cancer Database and Tissue Bank System (MOCDTBS) until use. Written informed consent was obtained from each participant before sample collection. cDNA from these samples were obtained from the MOCDTBS coordinated by Oral Cancer Research and Coordinating Centre (OCRCC), Faculty of Dentistry, were generated from RNA extracted from tissue specimens with ≥70% tumour or normal epithelial cells and only RNA with RNA integrity number (RIN) of more than 7 was used for cDNA synthesis.
For CTHRC1 protein expression determination and analysis, 74 OSCC formalin-fixed paraffin embedded (FFPE) tissues included were obtained from the archives of the Oral Pathology Diagnostic Laboratory, Faculty of Dentistry, University of Malaya consisting of specimens from surgical excisions of OSCC between 2003 to 2010. The normal oral mucosal tissues consisting of 9 FFPE tissues from alveolar mucosa obtained during removal of impacted wisdom tooth from individuals without any cancer or potentially malignant lesions. Tissue Micro Array (TMAs) was constructed which comprised of 1.0 mm core size of the FFPE tissues from OSCC and normal oral mucosa. The TMAs was constructed as previously described [25] by semi-automatic Tissue Arrayer Minicore (Alphelys, SAS, France). The representative areas of OSCC on the donor blocks were selected by 2 oral pathologists independently for the construction of the tissue microarray (TMA). Approximately three to six cores from the selected areas of donor blocks were then transferred into a recipient paraffin block. The TMAs were then sectioned at 4-µm for immunohistochemistry (IHC) and haematoxylin and eosin (H&E) staining. Socio-demographic, clinico-pathological and follow-up data of each sample were also obtained from the MOCDTBS. Survival information was available for all 74 OSCC patients, and the range of follow-up was 3-93 months, with mean and median overall survival of 32 months and 21 months respectively. This study was approved by the Medical Ethics Committee, Faculty of Dentistry, University of Malaya [Institutional Review Board Reference Number: DF OS1303/0003(P)].
Quantitative polymerase chain reaction (q-PCR)
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany) and cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instruction. CTHRC1 gene expression at the mRNA level was examined by quantitative polymerase chain reaction (q-PCR) using Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). All samples were run in triplicates in a total volume of 10 µl, which contains CTHRC1 Taqman probe (Hs00298917_m1), TaqMan® Fast Advanced Master Mix, nuclease free water, and 1 µl of cDNA template. Amplification was carried out with polymerase activation at 95ºC for 20 sec followed by 50 cycles at 95ºC for 3 sec and 60ºC for 30 sec. Human GAPDH was used as the reference gene. The relative expression analysis was performed using the 2-ΔΔCt relative quantification (RQ) method by comparing Ct values of CTHRC1 mRNA expression relative to the reference gene GAPDH, using 7500 Software v2.0.1 (Applied Biosystems, Foster City, CA, USA). Expressions of CTHRC1 in all OSCC samples were normalized to those from normal oral mucosa tissues using 7500 Software v2.0.1 (Applied Biosystems, Foster City, CA, USA). Expressions with fold change above 2 and below -2 were considered up- and down-regulated respectively.
Immunohistochemistry (IHC)
The protein expression of CTHRC1 was detected using immunohistochemistry. Immunohistochemistry was performed on 4µm thick TMA sections. In brief, TMA sections were deparaffinized with xylene and gradually rehydrated in descending grades of ethanol as described previously [26]. Antigen was retrieved by pressure cooking of the TMA section slides in 10 mM sodium citrate buffer (pH 6.0) at 20 psi, 121ºC for 30 sec and 90ºC for 10sec. Endogenous peroxidase activities of the TMA sections were blocked with endogenous peroxidase blocking agent (Dako, Kyoto, Japan) for 10 min in a humidified chamber followed by washing with phosphate buffered saline with 0.05% Tween-20 (PBST). The TMA sections were then incubated with 1:400 dilution of rabbit polyclonal primary antibody against CTHRC1 (Abcam, ab85739, MA, USA) [27] at room temperature for 45 min continued by washing with PBST. Then, TMA sections were treated with secondary antibodies for 30 min by Dako REALTM EnvisionTM Detection System, Peroxidase/DAB+, Rabbit/Mouse kit (Dako, Kyoto, Japan). The colour was developed by the addition of 3,3'-diaminobenzidine tetrahydrochloride (DAB) reagent for 5 min. Finally, TMA tissue sections were counterstained with haematoxylin, dehydrated through alcohols and xylene and mounted with DPX mountant (Fluka, Steinheim Germany). Primary melanoma tissue was used as positive control for CTHRC1 [12]. Negative control was performed using the same condition; except that the primary antibody was replaced with PBS on OSCC section that known to stain positively with CTHRC1 primary antibody.
Immunostaining assessment
All the digitalized immunostained tissue array slides were evaluated by two independent, experienced oral pathologists using the TMA software module 1.15.2 (3DHISTECH, Budapest, Hungary). Immunostaining was scored semi-quantitatively following the Immunohistochemical Scoring System [29-32]. IHC scoring was graded based on the proportion of staining intensity on a scale of negative to strong as follows: 0 = absent, +1 = low positive stained, +2 = moderate stained, and +3 = high positive stained cells. Percentage of positively stained cells was also recorded as follows: 0 = negative; 1= 1-10%; 2= 11-50%; 3= 51-80% and 4= 81%-100% of positive cells. The final immunohistochemical scoring was calculated by multiplying the intensity and percentage positive scores to obtain an immunoreactive score ranging from 0-12 [29, 30, 32]. The mean of immune-reactive scores evaluated by both oral pathologists were recorded and for those non-concordance cases, a consensus was reached by joint review of the each immunostained tissues to consolidate the final immunoreactive score to avoid any discrepancy.
Statistical analysis
All statistical analyses were analysed using the statistical software package SPSS 12.0.1 for Windows (SPSS Inc., Chicago, IL). The differentially expressed CTHRC1 between OSCC and normal oral mucosa samples was analysed using non-parametric Mann-Whitney test.
Statistical analysis for CTHRC1 protein expression (IHC) by categorizing the mean of final immunoreactive score into 2 groups. The cut-off point for this grouping was derived based on the optimal sensitivity and specificity obtained from receiver operating characteristic (ROC) curve analysis (Table S1). According to the ROC analysis, a cut-off point of 11.45 was determined where the area under the curve (AUC) was 0.703, and the sensitivity and specificity was 73.7% and 75.0% respectively. The immunoreactive mean score ≥ 11.45 and < 11.45 were categorized as high and low CTHRC1 expression respectively.
The associations between the expression of CTHRC1 and selected clinico-pathological parameters were analysed using univariate and multivariate logistic regression. Survival curves were plotted using the Kaplan-Meier analysis to correlate survival with CTHRC1 expression and the survival probability differences were compared by log rank tests. Cox regression analysis was conducted to determine the correlation between OSCC patients' survival and CTHRC1 protein expression after adjustment with confounder factors associated with OSCC. The p-value <0.05 was considered statistically significant.
Results
CTHRC1 mRNA expression
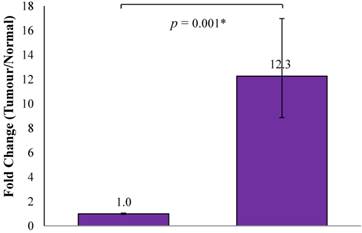
The current study showed that CTHRC1 mRNA up-regulation was seen in 38/45 (84.4%) of OSCCs, while 7/45 (15.6%) OSCC samples showed similar mRNA expression levels as normal oral mucosa samples (n=5) and none of OSCC samples showed down-regulation of CTHRC1 mRNA compared to normal oral mucosa samples. On average, the OSCC samples showed a 12.3-fold higher expression than normal oral mucosa samples, and this difference was statistically significant (p=0.001; Figure 1).
Average fold change of CTHRC1 in OSCC normalized with normal oral mucosa. On average, the mRNA expression level of CTHRC1 in OSCC samples was 12.3-fold higher expressed than that of normal oral mucosa samples (p=0.001).
CTHRC1 protein expression
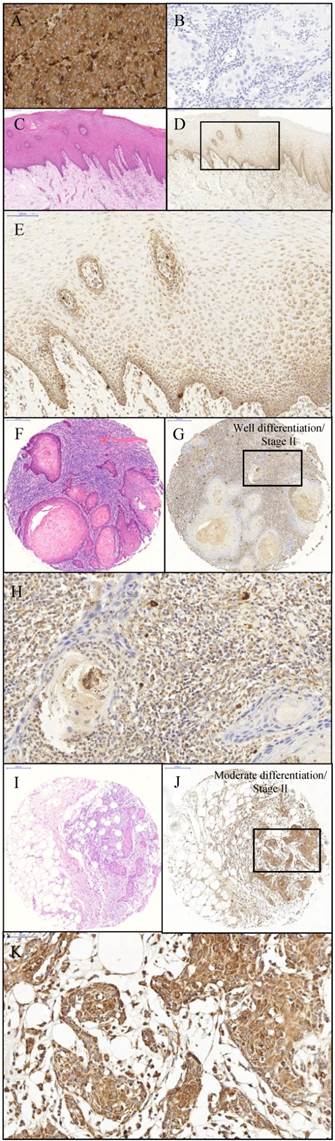
In the current study, protein expression of CTHRC1 was examined by immunohistochemical analysis. Primary melanoma tissue was used as the positive control for immunohistochemistry of CTHRC1 protein expression. The positive control tissue demonstrated strong granular cytoplasmic staining and negative nuclear staining (Figure 2A). Moreover, the negative control (OSCC tissue) for immunohistochemistry showed an absence of cytoplasmic and nuclear staining. (Figure 2B). IHC staining of CTHRC1 on a total of 9 normal oral mucosa showed a positive staining at the basement membrane but weak to negative cytoplasmic staining towards the spinous and keratinized layer (Figure 2D-E) in 8/9 (88.9%) samples. IHC staining on a total of 74 OSCC tissues demonstrated that 50.0% (37/74) of OSCCs exhibited a strong granular cytoplasmic staining in the epithelial tumour cells (Figure 2J-K).
Association of CTHRC1 protein expression levels with socio-demographic and clinico-pathological parameters
Table 1 shows the distribution of selected socio-demographic and clinico-pathological parameters and its association with CTHRC1 protein expression. Over-expression of CTHRC1 protein was found to be significantly associated with advanced stage pTNM staging (p = 0.010), positive lymph node metastasis (LNM)(p = 0.034), and tumour size ≥ 4cm (p = 0.011). However, only positive LNM remained significant in the multivariate binary logistic analyses (p = 0.044) even after adjustment for confounding factors such as age, gender, risk habits, tumour size, tumour site, pathological tumour-node-metastasis (pTNM) staging, lymph node metastasis, and Broder's grading (Table 2).
Immunohistochemistry of CHTRC1. The positive control: primary melanoma tissue (A) demonstrating expression of CTHRC1 protein. Strong cytoplasmic staining and negative nuclear staining was observed within primary melanoma tissue with magnification 1600x. The negative control: OSCC tissue (B) demonstrating lack of staining when primary antibody is omitted. The negative control showed the negative cytoplasmic and nuclear staining within OSCC tissue with magnification 1600x. Normal oral mucosa (C) H&E stain (Original magnification 400x); (D) anti-CTHRC1 (Original magnification 400x) (E) Expression of CTHRC1: moderate cytoplasmic staining was observed within the epithelial cells of the normal oral mucosa (Magnification 800x). OSCC (F and I) H&E stain (Magnification 400x); (G and H) Expression of CTHRC1: moderate cytoplasmic staining and negative nuclear staining was observed within OSCC tumour cells (G) Magnification 400x; (H) Original magnification 1600x; (J and K) Strong cytoplasmic staining and negative nuclear staining was observed within OSCC tumour cells (J) Magnification 400x; (K) Original magnification 1600x.
Socio-demographic and clinico-pathological characteristics for 74 OSCC patients included in the IHC analysis.
| Variables | Subgroups | Total (n) | Percentage (%) |
|---|---|---|---|
| Gender | Male | 29 | 39.19 |
| Female | 45 | 60.81 | |
| Age (years) | <45 | 6 | 8.11 |
| ≥45 | 68 | 91.89 | |
| Betel quid chewing* | No | 34 | 45.95 |
| Yes | 39 | 52.70 | |
| Smoking* | No | 58 | 78.38 |
| Yes | 15 | 20.27 | |
| Drinking* | No | 47 | 63.51 |
| Yes | 26 | 35.14 | |
| pTNM Staging | Early stage | 29 | 39.19 |
| Advanced stage | 45 | 60.81 | |
| Lymph node | Negative | 45 | 60.81 |
| Positive | 29 | 39.19 | |
| Tumour size | < 4cm | 43 | 58.11 |
| ≥ 4cm | 31 | 41.89 | |
| Tumour site | Buccal mucosa | 27 | 36.49 |
| Gum | 11 | 14.86 | |
| Tongue | 25 | 33.78 | |
| Other ** | 11 | 14.86 | |
| Broder's grading | Well | 29 | 39.19 |
| Moderate or poor | 45 | 60.81 | |
| Overall Survival (Month) | Range (3-93) | 74 | 100 |
| Mean (32.22) | |||
| Median (21) |
*one data missing.
**Other = Lip and floor of mouth.
Protein expression of CTHRC1 in correlation with survival status
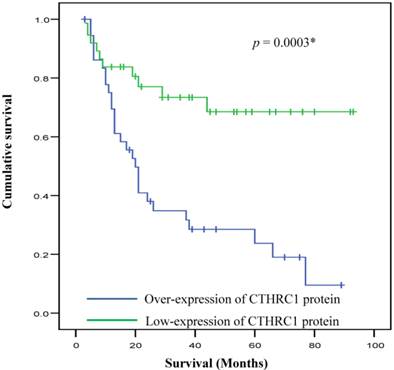
Kaplan-Meier survival analysis demonstrated that survival of OSCC patients was significantly different between patient with low and high expression of CTHRC1 protein. Patients with high expression of CTHRC1 was associated with significantly poorer survival outcome as compared to low-expression (p = 0.0003) (Figure 3). Multivariate Cox regression analysis revealed that CTHRC1 protein expression level is an independent prognostic factor for OSCC. A significant 2.6-fold increased risk of mortality (HRR 2.59, p=0.035) was seen for high CTHRC1 expression even after adjustment for factors associated with OSCC such as age, gender, risk habits, tumour size, tumour site, pTNM staging, lymph node metastasis and Broder's grading (Table 3).
Discussion
Based on our previous array CGH study, 8q11.1-q24.4 is one of the most frequently amplified CNAs regions where the copy number gain (54%) in the OSCC tissues [16]. In this current study, CTHRC1 located at chromosome 8q22.3 was analysed to determine the mRNA expression levels of CTHRC1 in OSCC and normal oral mucosa samples using quantitative-PCR technique. CTHRC1 was indeed found to be over-expressed at the transcript level in OSCC compared to normal oral mucosa samples (12.3 fold change, p = 0.001). According to the available published data in cancer for CTHRC1, the current finding is in concordance with other studies as confirmed by q-PCR, which also showed that CTHRC1 was consistently highly expressed (≈ 8 fold change) in various types of human cancers including metastatic melanoma [12], colorectal cancer [33], breast cancer [34], and pancreatic ductal adenocarcinomas (PADC) [18] compared to their respective normal samples.
Overall survival curves for OSCC in relation to CTHRC1 protein expression. Kaplan-Meier analyses with log-rank test indicated that high CTHRC1 expression signature had significantly lower 5-year overall survival rate compared to low CTHRC1 expression in OSCC patients (p=0.0003*).
Association between CTHRC1 protein expression with socio-demographic and clinico-pathological characteristics of OSCC patients.
| Subgroups | CTHRC1 protein expression (n, %) | Univariate logistic regression | Multivariate logistic regressiona | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables | Low | High | OR | 95% CI | p value | OR | 95% CI | p value | |
| Gender | Male | 15 (40.5) | 14 (37.8) | 1 | (0.35, 2.27) | 0.812 | 1 | (0.59, 12.97) | 0.196 |
| Female | 22 (59.5) | 23 (62.2) | 0.89 | 2.77 | |||||
| Age (years) | <45 | 5 (13.5) | 1 (2.7) | 1 | (0.62, 50.73) | 0.124 | 1 | (0.66, 375.17) | 0.089 |
| ≥45 | 32 (86.5) | 36 (97.3) | 5.63 | 15.71 | |||||
| Betel quid chewing | No | 19 (52.8) | 15 (40.5) | 1 | (0.65, 4.14) | 0.296 | 1 | (0.28, 4.52) | 0.871 |
| Yes | 17 (47.2) | 22 (59.5) | 1.64 | 1.12 | |||||
| Smoking | No | 28 (77.8) | 30 (81.1) | 1 | (0.26, 2.55) | 0.727 | 1 | (0.20, 8.08) | 0.798 |
| Yes | 8 (22.2) | 7 (18.9) | 0.82 | 1.27 | |||||
| Drinking | No | 21 (58.3) | 26 (70.3) | 1 | (0.23, 1.56) | 0.289 | 1 | (0.09, 1.23) | 0.098 |
| Yes | 15 (41.7) | 11 (29.7) | 0.59 | 0.33 | |||||
| pTNM Staging | Early stage | 20 (54.1) | 9 (24.3) | 1 | (1.36, 9.86) | 0.010* | 1 | (0.03, 3.29) | 0.331 |
| Advanced stage | 17 (45.9) | 28 (75.7) | 3.66 | 0.31 | |||||
| Lymph node | Negative | 27 (73.0) | 18 (48.6) | 1 | (1.08, 7.52) | 0.034* | 1 | (1.06, 51.36) | 0.044* |
| Positive | 10 (27.0) | 19 (51.4) | 2.85 | 7.37 | |||||
| Tumour size | < 4cm | 27 (73.0) | 16 (43.2) | 1 | (1.34, 9.39) | 0.011* | 1 | (0.94, 43.51) | 0.058 |
| ≥ 4cm | 10 (27.0) | 21 (56.8) | 3.54 | 6.38 | |||||
| Tumour site | Buccal mucosa | 11 (29.7) | 16 (43.2) | 1 | (0.48, 1.11) | 0.144 | 1 | (0.45, 1.30) | 0.32 |
| Gum | 4 (10.8) | 7 (18.9) | 0.73 | 0.76 | |||||
| Tongue | 16 (43.2) | 9 (24.3) | |||||||
| Other** | 6 (16.2) | 5 (13.5) | |||||||
| Broder's grading | Well | 15 (40.5) | 14 (37.8) | 1 | (0.35, 2.27) | 0.812 | 1 | (0.34, 3.59) | 0.870 |
| Moderate or Poor | 22 (59.5) | 23 (62.2) | 0.89 | 1.10 | |||||
**Other = Lip and floor of mouth.
aAdjusted for gender, age, risk habits, pTNM staging, lymph node, tumour size, tumour site and Broder's grading. OR represented odd ratio.
* Indicated significant p- value (p < 0.05).
Multivariate Cox regression analysis of overall survival.
| Variables | Category | Multivariate Cox regressiona | ||
|---|---|---|---|---|
| HRR | 95% CI | p value | ||
| CTHRC1 protein expression | Low | 1.00 | (1.07, 6.25) | 0.035* |
| High | 2.59 | |||
| Age group | <45 | 1.00 | (0.13, 2.14) | 0.377 |
| ≥45 | 0.54 | |||
| Gender | Male | 1.00 | (0.37, 3.00) | 0.917 |
| Female | 1.06 | |||
| Chewing | No | 1.00 | (0.20, 1.30) | 0.158 |
| Yes | 0.51 | |||
| Smoking | No | 1.00 | (0.11, 1.70) | 0.225 |
| Yes | 0.42 | |||
| Drinking | No | 1.00 | (0.30, 1.60) | 0.393 |
| Yes | 0.69 | |||
| Tumour size | < 4cm | 1.00 | (0.49, 3.60) | 0.582 |
| ≥ 4cm | 1.32 | |||
| Lymph node | Negative | 1.00 | (1.13, 9.86) | 0.029* |
| Positive | 3.34 | |||
| pTMN Staging | Early | 1.00 | (0.26, 4.16) | 0.955 |
| Advanced | 1.04 | |||
| Tumour site | Buccal mucosa | 1.00 | (0.65, 1.32) | 0.675 |
| Gum | 0.93 | |||
| Tongue | ||||
| Other* | ||||
| Broder's grading | Well | 1.00 | (0.83, 3.92) | 0.135 |
| Poor | 1.80 | |||
**Other = Lip and floor of mouth
aAdjusted for gender, age, risk habits, pTNM staging, lymph node, tumour size, tumour site and Broder's grading. HRR represented hazard rate ratio
* Indicated significant p- value (p < 0.05)
This is the first study to report the association between CTHRC1 expression levels and clinico-pathological parameters of OSCC patients. CTHRC1 protein expression levels in a total of 74 OSCC and 9 normal oral mucosa samples were determined by IHC assay. In this study, over-expression of CTHRC1 protein was found to be significantly associated with positive LNM, tumour size ≥ 4cm and advanced stage pTNM staging, in patients with OSCC. However, only the positive LNM remained significant after adjustment with other confounders such as age, gender, risk habits, tumour size, tumour site, pTNM staging, and Broder's grading in multivariate logistic regression analysis. This finding suggested that CTHRC1 expression level could be a potential independent biomarker in predicting lymph node status of OSCC patients. This observation can be further explained in the study by Liu et al. (2013) which demonstrated that CTHRC1 is up-regulated by DPAGT1 and canonical Wnt signalling via the N-Glycosylation mechanism in OSCC. This enables to promote cancer cell migration and invasion in oral carcinogenesis [21]. In addition, Park et al. (2013) suggested that the up regulation of CTHRC1 in pancreatic cancer might be one of the causal effects that leads to tumour metastasis by activate several key signalling molecules, including Src, focal adhesion kinase, paxillin, mitogen-activated protein kinase, extracellular signal-regulated kinase and Rac1 [18]. Activation of these signalling molecules would dysregulate the tumour cell migration and adhesion activities and lead to tumour cell metastasis. The overall survival of OSCC patients in this study was found to be associated with CTHRC1 expression levels. Patients with high CTHRC1 expression were found to have poorer prognosis than that of patients with low CTHRC1 expression. The 3 and 5-years overall survival of patients with high CTHRC1 expression level were only 31.7% and 23.7% respectively, while the corresponding survival rate for patients with low CTHRC1 expression were approximately 73.4% and 68.5%, respectively. This indicates that OSCC patients with high CTHRC1 expression had more than 2 fold increased risk for poorer survival outcome compared to patients with low expression. This observation is in concordance with a recent study on gastric and colorectal cancer which also reported that prognosis of patients with low CTHRC1 expression was better than that of patients displaying high expression [33, 35].
As the over-expression of CTHRC1 protein was significantly correlated with lymph node metastasis in OSCC patients and the lymph node metastasis is a major determination of survival outcome, therefore, this factor was taken into consideration in the multivariate Cox regression analysis. Notably, CTHRC1 expression levels remained a significant predictive factor for patients' survival (p = 0.035) after adjustment with other major factors associated with OSCC survival including lymph node status. This indicated that CTHRC1 protein expression is an independent prognostic predictor for overall survival among OSCC patients, suggesting that CTHRC1 can affect survival independent of lymph node metastasis. Therefore, over-expression of CTHRC1 protein expression could have lymph node metastasis dependent and independent roles that lead to poorer survival among patients who over-express this protein in their tumours.
Although there are several limitations that may weaken the generalization of current finding including only involved a relatively small amount of specimens that are available and has not shown the mechanism of CTHRC1 protein driver of malignancy. However, current study still able to demonstrate high expression of CTHRC1 protein shown significant correlations with positive lymph node metastasis. Moreover, current finding also indicated that CTHRC1 expression level can be a major prognosis indicator for predict survival outcome of OSCC patients.
Conclusion
In conclusion, the present study revealed that CTHRC1 was over-expressed in OSCCs compared to normal controls. Over-expression of this protein could be an independent predictor for nodal metastasis. This suggests that CTHRC1 expression signature could assist clinicians to improve the assessment of predicting the cervical lymph node metastasis status of oral cancer patients. Furthermore, the current data also suggests that low CTHRC1 expression level is a significant good prognostic marker for OSCC. Future studies are required to further validate and investigate the functional roles of this gene which could subsequently lead to the development of novel therapeutic target in the treatment of oral cancer patients.
Supplementary Material
Table S1.
Acknowledgements
This study was supported by University of Malaya Research Grant (RP002E-13HTM) and Ministry of Higher Education (MOHE) Malaysia High Impact Research grant with project number: UM.C/625/1/HIR/MOHE/DENT/06. The authors acknowledged the Oral Cancer Research and Coordinating Centre (OCRCC), Faculty of Dentistry, University of Malaya (UM) for providing tissue and data from the Malaysian Oral Cancer Database & Tissue Bank System (MOCDTBS). The authors also thanked Professor Yi-Hsin Yang, a biostatistician from Kaohsiung Medical University, Taiwan and the clinicians and pathologists from Oral Health Division, Ministry of Health Malaysia for their expertise.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69-90 doi:10.3322/caac.20107
2. Sankaranarayanan R. Oral cancer in India: an epidemiologic and clinical review. Oral surgery, oral medicine, and oral pathology. 1990;69:325-30
3. Lau SH, Jalil AA, Zain RB. Cancer Incidence in Peninsular Malaysia 2003-2005: The Third Report of the National Cancer Registry, Ministry of Health; 2008.
4. Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C. et al. Cancer statistics, 2006. CA: a cancer journal for clinicians. 2006;56:106-30
5. Dalirsani Z, Farajnia S, Javadzadeh Y, Mehdipour M, Koozegari S. The Effects of 5-fluorouracil Alone and in Combination with 13-cis Retinoic Acid and Vitamin D3 on Human Oral Squamous Cell Carcinoma Lines. The journal of contemporary dental practice. 2012;13:345-50
6. Sun P, Gao J, Liu YL, Wei LW, Wu LP, Liu ZY. RNA interference (RNAi)-mediated vascular endothelial growth factor-C (VEGF-C) reduction interferes with lymphangiogenesis and enhances epirubicin sensitivity of breast cancer cells. Molecular and cellular biochemistry. 2008;308:161-8 doi:10.1007/s11010-007-9624-1
7. Saraswathi TR, Kavitha B, Vijayashree Priyadharsini J. Gene therapy for oral squamous cell carcinoma: an overview. Indian journal of dental research: official publication of Indian Society for Dental Research. 2007;18:120-3
8. Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer metastasis reviews. 2007;26:645-62 doi:10.1007/s10555-007-9082-y
9. Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nature reviews Cancer. 2005;5:845-56 doi:10.1038/nrc1739
10. Durmus T, LeClair RJ, Park KS, Terzic A, Yoon JK, Lindner V. Expression analysis of the novel gene collagen triple helix repeat containing-1 (Cthrc1). Gene expression patterns: GEP. 2006;6:935-40
11. Pyagay P, Heroult M, Wang Q, Lehnert W, Belden J, Liaw L. et al. Collagen triple helix repeat containing 1, a novel secreted protein in injured and diseased arteries, inhibits collagen expression and promotes cell migration. Circulation research. 2005;96:261-8
12. Tang L, Dai DL, Su M, Martinka M, Li G, Zhou Y. Aberrant expression of collagen triple helix repeat containing 1 in human solid cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:3716-22
13. Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860-7
14. Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324-31
15. Li J, Zhang YP, Kirsner RS. Angiogenesis in wound repair: angiogenic growth factors and the extracellular matrix. Microscopy research and technique. 2003;60:107-14 doi:10.1002/jemt.10249
16. Vincent-Chong VK, Anwar A, Karen-Ng LP, Cheong SC, Yang YH, Pradeep PJ. et al. Genome wide analysis of chromosomal alterations in oral squamous cell carcinomas revealed over expression of MGAM and ADAM9. PloS one. 2013;8:e54705. doi:10.1371/journal.pone.0054705
17. Tameda M, Sugimoto K, Shiraki K, Yamamoto N, Okamoto R, Usui M. et al. Collagen triple helix repeat containing 1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation and motility. International journal of oncology. 2014;45:541-8 doi:10.3892/ijo.2014.2445
18. Park EH, Kim S, Jo JY, Kim SJ, Hwang Y, Kim JM. et al. Collagen triple helix repeat containing-1 promotes pancreatic cancer progression by regulating migration and adhesion of tumor cells. Carcinogenesis. 2013;34:694-702 doi:10.1093/carcin/bgs378
19. Wang P, Wang YC, Chen XY, Shen ZY, Cao H, Zhang YJ. et al. CTHRC1 is upregulated by promoter demethylation and transforming growth factor-beta1 and may be associated with metastasis in human gastric cancer. Cancer science. 2012 doi:10.1111/j.1349-7006.2012.02292.x
20. Shibamoto S, Higano K, Takada R, Ito F, Takeichi M, Takada S. Cytoskeletal reorganization by soluble Wnt-3a protein signalling. Genes to cells: devoted to molecular & cellular mechanisms. 1998;3:659-70
21. Liu G, Sengupta PK, Jamal B, Yang HY, Bouchie MP, Lindner V. et al. N-glycosylation induces the CTHRC1 protein and drives oral cancer cell migration. The Journal of biological chemistry. 2013;288:20217-27 doi:10.1074/jbc.M113.473785
22. Chen YL, Wang TH, Hsu HC, Yuan RH, Jeng YM. Overexpression of CTHRC1 in hepatocellular carcinoma promotes tumor invasion and predicts poor prognosis. PloS one. 2013;8:e70324. doi:10.1371/journal.pone.0070324
23. Kalnins IK, Leonard AG, Sako K, Razack MS, Shedd DP. Correlation between prognosis and degree of lymph node involvement in carcinoma of the oral cavity. American journal of surgery. 1977;134:450-4
24. Woolgar JA, Scott J, Vaughan ED, Brown JS, West CR, Rogers S. Survival, metastasis and recurrence of oral cancer in relation to pathological features. Annals of the Royal College of Surgeons of England. 1995;77:325-31
25. Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S. et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature medicine. 1998;4:844-7
26. Zanaruddin SN, Saleh A, Yang YH, Hamid S, Mustafa WM, Khairul Bariah AA. et al. Four-protein signature accurately predicts lymph node metastasis and survival in oral squamous cell carcinoma. Human pathology. 2013;44:417-26 doi:10.1016/j.humpath.2012.06.007
27. Kim HC, Kim YS, Oh HW, Kim K, Oh SS, Kim JT. et al. Collagen triple helix repeat containing 1 (CTHRC1) acts via ERK-dependent induction of MMP9 to promote invasion of colorectal cancer cells. Oncotarget. 2014;5:519-29
28. Sapkota A, Hsu CC, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Mates D. et al. Dietary risk factors for squamous cell carcinoma of the upper aerodigestive tract in central and eastern Europe. Cancer causes & control: CCC. 2008;19:1161-70 doi:10.1007/s10552-008-9183-0
29. Kamoi S, AlJuboury MI, Akin MR, Silverberg SG. Immunohistochemical staining in the distinction between primary endometrial and endocervical adenocarcinomas: another viewpoint. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists. 2002;21:217-23
30. Koo CL, Kok LF, Lee MY, Wu TS, Cheng YW, Hsu JD. et al. Scoring mechanisms of p16INK4a immunohistochemistry based on either independent nucleic stain or mixed cytoplasmic with nucleic expression can significantly signal to distinguish between endocervical and endometrial adenocarcinomas in a tissue microarray study. Journal of translational medicine. 2009;7:25. doi:10.1186/1479-5876-7-25
31. Khoury T, Tan D, Wang J, Intengan M, Yang J, Alrawi S. et al. Inclusion of MUC1 (Ma695) in a panel of immunohistochemical markers is useful for distinguishing between endocervical and endometrial mucinous adenocarcinoma. BMC clinical pathology. 2006;6:1. doi:10.1186/1472-6890-6-1
32. Liao CL, Lee MY, Tyan YS, Kok LF, Wu TS, Koo CL. et al. Progesterone receptor does not improve the performance and test effectiveness of the conventional 3-marker panel, consisting of estrogen receptor, vimentin and carcinoembryonic antigen in distinguishing between primary endocervical and endometrial adenocarcinomas in a tissue microarray extension study. Journal of translational medicine. 2009;7:37. doi:10.1186/1479-5876-7-37
33. Tan F, Liu F, Liu H, Hu Y, Liu D, Li G. CTHRC1 is associated with peritoneal carcinomatosis in colorectal cancer: a new predictor for prognosis. Medical oncology. 2013;30:473. doi:10.1007/s12032-013-0473-3
34. Kim JH, Baek TH, Yim HS, Kim KH, Jeong SH, Kang HB. et al. Collagen Triple Helix Repeat Containing-1 (CTHRC1) Expression in Invasive Ductal Carcinoma of the Breast: The Impact on Prognosis and Correlation to Clinicopathologic Features. Pathology oncology research: POR. 2013 doi:10.1007/s12253-013-9636-y
35. Gu L, Liu L, Zhong L, Bai Y, Sui H, Wei X. et al. Cthrc1 overexpression is an independent prognostic marker in gastric cancer. Human pathology. 2014;45:1031-8 doi:10.1016/j.humpath.2013.12.020
Author contact
![]() Corresponding author: Rosnah Binti Zain, Oral Cancer Research and Coordinating Centre (OCRCC), Faculty of Dentistry, University of Malaya, 50603 Kuala Lumpur, Malaysia. Tel: +60379674896 Fax: +60379547301 E-mail address: rosnahbz12com or rosnahmzedu.my.
Corresponding author: Rosnah Binti Zain, Oral Cancer Research and Coordinating Centre (OCRCC), Faculty of Dentistry, University of Malaya, 50603 Kuala Lumpur, Malaysia. Tel: +60379674896 Fax: +60379547301 E-mail address: rosnahbz12com or rosnahmzedu.my.

 Global reach, higher impact
Global reach, higher impact