Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(6):458-467. doi:10.7150/ijms.11808 This issue Cite
Research Paper
Altered Activity and Expression of Cytosolic Peptidases in Colorectal Cancer
1. Department of Nursing I, School of Nursing, University of the Basque Country (UPV/EHU), Leioa, Bizkaia, Spain
2. Department of Physiology, Faculty of Medicine and Dentistry, University of the Basque Country (UPV/EHU), Leioa, Bizkaia, Spain
3. Department of Surgery, Basurto University Hospital, University of the Basque Country (UPV/EHU), Bilbao, Bizkaia, Spain
4. Institute for Chemical Research, CSIC-Isla de la Cartuja, Sevilla, Spain;
5. Department of Anatomic Pathology, Cruces University Hospital, University of the Basque Country (UPV/EHU), Barakaldo, Bizkaia, Spain
6. BioCruces Health Research Institute, Barakaldo, Bizkaia, Spain
Received 2015-2-6; Accepted 2015-4-29; Published 2015-6-2
Abstract
Background and Objective: The role of peptidases in carcinogenic processes and their potential usefulness as tumor markers in colorectal cancer (CRC) have been classically attributed to cell-surface enzymes. The objective of the present study was to analyze the activity and mRNA expression of three cytosolic peptidases in the CRC and to correlate the obtained results with classic histopathological parameters for tumor prognosis and survival.
Methods: The activity and mRNA levels of puromycin-sensitive aminopeptidase (PSA), aminopeptidase B (APB) and pyroglutamyl-peptidase I (PGI) were measured by fluorimetric and quantitative RT-PCR methods in colorectal mucosa and tumor tissues and plasma samples from CRC patients (n=81).
Results: 1) PSA and APB activity was higher in adenomas and carcinomas than in the uninvolved mucosa. 2) mRNA levels of PSA and PGI was lower in tumors. 3) PGI activity in CRC tissue correlated negatively with histological grade, tumor size and 5-year overall suvival of CRC patients. 4) Higher plasmatic APB activity was independently associated with better 5-year overall survival.
Conclusions: Data suggest that cytosolic peptidases may be involved in colorectal carcinogenesis and point to the determination of this enzymes as a valuable method in the determination of CRC prognosis.
Keywords: colorectal cancer, prognosis, survival, cytosolic, soluble, peptidases.
Introduction
Colorectal cancer (CRC) is the second commonest malignancy in males and females in Europe [1] and the third in the United States [2]. Huge resources are being invested in prevention and early diagnosis of this disease. Population-based screening campaigns try to discover as much early tumors and precursor lesions as possible, aiming to decrease the incidence of the disease, to simplify the clinical management of patients once the lesion develops, and to improve survival [3]. However, despite of the advances in screening, surgery and radio and chemotherapy, it is still the second cause of cancer deaths in the developed countries [4].
The molecular classification of CRC proposed by Jass in 2007 [5] has been of much help in the understanding of the diverse etiopathogenetic mechanisms of this disease. From a pathological perspective, adenomatous lesions in the large bowel are fully accepted precursors of CRC and the adenoma-carcinoma sequence still provides a solid model for research on carcinogenesis [4,6]. These adenomatous polyps and subsequent carcinoma appear to develop with sequential genetic alterations which affect to the expression of several proteins, such as peptidases [7,8].
Peptidases play a key role in cell proliferation, angiogenesis and tumor invasiveness by regulating extracellular matrix degradation and cell signalling and recognition [8,9]. The expression and activity of these enzymes vary in cancer depending on the several clinicopathological parameters like histological grade, stage, and patient survival [8-11]. For this reason, peptidases are useful tools in the development of clinical strategies for treatment and follow up of cancer patients.
Although the role of these proteins in carcinogenic processes and their potential usefulness as tumor markers in CRC was initially proposed for cell-surface enzymes [8,12,13], it has been demonstrated that cytosolic peptidases also participate in several hallmarks of cancer [14-19]. In addition, it has been reported that the activity of different cytosolic peptidases such as puromycin-sensitive aminopeptidase (PSA or EC 3.4.11.14), aminopeptidase B (APB or EC 3.4.11.6) and pyroglutamyl-peptidase I (PGI or EC 3.4.19.3) was altered in breast [19,20], kidney [21,22] and thyroid tumors [23].
Regarding CRC, recently we demonstrated that the activity and mRNA expression of prolyl endopeptidase (PEP) and aspartyl aminopeptidase (ASP), was altered throughout the uninvolved colorectal mucosa-adenoma-carcinoma sequence. Moreover, tissue and plasmatic activity of both enzymes was correlated with tumor invasiveness and 5-year survival of CRC patients [16-18]. All these evidences point to the analysis of cytosolic peptidases as a promising tool in the design of new diagnostic/prognostic biomarkers in CRC.
In this context, the objective of the present work was to study the activity and mRNA expression of PSA, APB and PGI in the adenoma-adenocarcinoma sequence and plasma samples from patients with CRC, and to correlate the obtained results with classic histopathological parameters for tumor prognosis and survival.
Materials & Methods
The authors declare that all the experiments carried out in this study comply with current Spanish laws and conform to the principles outlined in the Declaration of Helsinki.
Patients
81 patients with CRC were prospectively included in the study. Males predominated in the series (51M/30F), the average age being 70.1 years for males and 67 years for females. Mean follow-up was 50.2 months (range 3-80). All patients received partial colectomies. In 16 of these patients, both adenomatous polyps and adenocarcinomas were diagnosed. These cases were used to analyze peptidase activity and mRNA levels throughout the uninvolved mucosa-adenoma-adenocarcinoma sequence.
Follow-up was closed by December 31, 2012. At that time, 25 patients had died of disease. AJCC system [24] has been applied to assign Stage and Grade. Clinical data included in the study were retrieved from the patient clinical records and are summarized in Table 1A. Plasma was also collected preoperatively and analyzed in 40 of these patients. Table 1B shows the clinicopathological data in this subset of patients. Plasma from 28 healthy volunteers with no clinical history of neoplastic diseases was used as control sample.
Tissue Specimens
Surgical resections were submitted in fresh to the Pathology Lab within a period of 30 minutes after removal. Handling of specimens was performed following conventional protocols for the management of surgical resections of colon and rectum [25]. Tumor characteristics were recognized on gross examination and selected fragments of tumor were frozen in isopentane and stored at -80ºC. Besides, peripheral venous blood samples from 40 of these patients were collected prior to surgery in EDTA tubes and centrifuged at 1500 rpm during 15 minutes. The obtained plasma was also stored at -80ºC. Enzyme assays have also been performed in plasma obtained from 28 healthy volunteers (matched by sex and age).
Sample preparation
Explanted tumor and non-tumor samples were homogenized in 10 mM Tris-HCl buffer at pH 7.4, using a Heidolph PZR 50 Selecta homogenizer, and ultracentrifuged in a Centrikon T-2070 Kontron Instruments apparatus at 100,000g for 35 minutes. The resulting supernatants (soluble fractions) were used to measure PGI, APB and PSA activity [19,26]. Previously collected plasma samples were used to determine plasmatic PGI, APB and PSA activity. All the above-described steps were carried out at 4 °C.
Peptidase activity in CRC tissue according to clinicopathological characteristics. Values are means ± SE of peptidase activity recorded as pmol of units of peptidase (UP) per milligram of protein.
| Variables | n = | APB | PSA | PGI | |||
|---|---|---|---|---|---|---|---|
| UP/mg prot (Mean ± S.E ) | Mann-Whitney (p =) | UP/mg prot (Mean ± S.E ) | Mann-Whitney (p =) | UP/mg prot (Mean ± S.E ) | Mann-Whitney (p =) | ||
| Grade | |||||||
| Low (G1-G2) | 64 | 13946 ± 1104 | 0.101 | 15570 ± 1164 | 0.385 | 186.84 ± 28.12 | 0.011 |
| High (G3-G4) | 17 | 12591 ± 4562 | 13058 ± 2994 | 126.25 ± 5.89 | |||
| Stage | |||||||
| Low (T1-T2) | 11 | 11474 ± 2471 | 0.847 | 13894 ± 2418 | 0.916 | 152.2 ±55.52 | 0.606 |
| High (T3-T4) | 70 | 14235 ± 1229 | 15501 ± 1214 | 183.96 ± 27.64 | |||
| Nodal invasion | |||||||
| No | 50 | 13429 ± 1371 | 0.625 | 14510 ± 1316 | 0.839 | 164.6 ± 18.09 | 0.820 |
| Yes | 31 | 14493 ± 1920 | 16810 ± 1865 | 209.33 ± 69.79 | |||
| Distant Metastases | |||||||
| No | 74 | 13757 ± 1177 | 0.589 | 14797 ± 1105 | 0.349 | 185.31 ± 26.72 | 0.183 |
| Yes | 7 | 13776 ± 3797 | 18917 ± 4143 | 119.33 ± 44.22 | |||
| Lymphatic invasion | |||||||
| No | 72 | 13606 ± 1179 | 0.333 | 15118 ± 1144 | 0.548 | 188.44 ± 27.74 | 0.050 |
| Yes | 9 | 14714 ± 3517 | 15884 ± 3580 | 116.25 ± 25.7 | |||
| Vascular invasion | |||||||
| No | 66 | 14479 ± 1166 | 0.161 | 15379 ± 1082 | 0.705 | 192.04 ± 29.9 | 0.429 |
| Yes | 15 | 11000 ± 2838 | 14629 ± 3401 | 126.5 ± 22.11 | |||
| Perineural invasion | |||||||
| No | 77 | 13677 ± 1139 | 0.861 | 15120 ± 1111 | 0.846 | 180.21 ± 25.33 | 0.949 |
| Yes | 4 | 12948 ± 2542 | 15584 ± 2019 | 154 ± 24.4 | |||
| Grouped stage | |||||||
| Low (0-IIC) | 46 | 13505 ± 1443 | 1.000 | 14332 ± 1375 | 0.542 | 162.84 ± 18.98 | 0.758 |
| High (IIIA-IV) | 35 | 14242 ± 1735 | 16918 ± 1671 | 208.2 ± 62.44 | |||
| Topographic distribution | |||||||
| Colon | 14064 ± 828 | 0.023 | 16108 ± 724 | 0.044 | 195 ± 18 | 0.026 | |
| Rectum | 10580 ± 1162 | 13009 ± 1142 | 130 ± 15 | ||||
Peptidase activity in plasma from CRC patients according to clinicopathological characteristics. Values are means ± SE of units of peptidase per liter of plasma (UP/L).
| Variables | n = | APB | PSA | PGI | |||
|---|---|---|---|---|---|---|---|
| UP/L (Mean ± S.E ) | Mann-Whitney (p =) | UP/L (Mean ± S.E ) | Mann-Whitney (p =) | UP/L (Mean ± S.E ) | Mann-Whitney (p =) | ||
| Grade | |||||||
| Low (G1-G2) | 33 | 100.16 ± 7.63 | 0.364 | 395.80 ± 38.71 | 0.064 | 25.24 ± 4.23 | 0.242 |
| High (G3-G4) | 7 | 88.75 ± 19.23 | 253.25 ± 43.82 | 8.49 ± 0.91 | |||
| Stage | |||||||
| Low (T1-T2) | 9 | 100.6 ± 24.84 | 0.915 | 360 ± 113 | 1.000 | 9.27 ± 1.37 | 0.421 |
| High (T3-T4) | 31 | 98.17 ± 7.05 | 379.5 ± 36.5 | 25.77 ± 4.38 | |||
| Nodal invasion | |||||||
| No | 29 | 98.9 ± 7.16 | 0.802 | 380.35 ± 45.65 | 0.918 | 29.74 ± 4.79 | 0.073 |
| Yes | 11 | 97.89 ± 16.8 | 366.78 ± 52.56 | 7.78 ± 0.65 | |||
| Distant Metastases | |||||||
| No | 37 | 97.23 ± 6.6 | 0.959 | 378.81 ± 38.22 | 0.700 | 23.57 ± 4.09 | 0.330 |
| Yes | 3 | 110.33 ± 42.99 | 353 ± 82.6 | 17.39 ± 11.14 | |||
| Lymphatic invasion | |||||||
| No | 34 | 95.8 ± 6.7 | 0.519 | 373 ± 39.42 | 1.000 | 25.39 ± 4.21 | 0.068 |
| Yes | 6 | 116 ± 30.92 | 395.75 ± 69.66 | 7.56 ± 1.31 | |||
| Vascular invasion | |||||||
| No | 33 | 101.96 ± 7.94 | 0.669 | 396.56 ± 41.03 | 0.557 | 23.49 ± 4.51 | 0.522 |
| Yes | 7 | 85.67 ± 14.91 | 297.83 ± 56.03 | 20.77 ± 6.83 | |||
| Perineural invasion | |||||||
| No | 38 | 98.93 ± 7.25 | 0.862 | 380.32 ± 35.97 | 0.829 | 22.26 ± 3.88 | 0.328 |
| Yes | 2 | 89 ± 6.1 | 259 ± 31.2 | 41.74 ± 8.23 | |||
| Grouped stage | |||||||
| Low (0-IIC) | 28 | 99.84 ± 7.48 | 0.740 | 388.47 ± 47.36 | 0.885 | 29.23 ± 5.02 | 0.136 |
| High (IIIA-IV) | 12 | 96.2 ± 15.12 | 352.7 ± 49.07 | 10.96 ± 3.23 | |||
PGI, APB, PSA activity measurements
PGI, APB and PSA activity was fluorimetrically measured using a modified version of the method described by Zambotti-Villela et al. [27]. PGI was quantified with pGlu-β-naphthylamide (0.125mM) as substrate and APB with Arg-β-naphthylamide (0.125mM). In the case of PSA was measured using Ala-β-naphthylamide (0.125mM) as substrate. To discriminate between PSA and aminopeptidase N forms of total alanine aminopeptidase activity, incubations with the specific PSA inhibitor puromycin (40µM) were performed in parallel.
The assay is based on the fluorescence of β-naphthylamine generated from the hydrolysis of the substrate by the enzyme. The components of the assays mixtures (1 ml) included the following: PGI assay [(50mM of sodium phosphate buffer (pH 7.4), 2 mM of DL-dithiothreitol and 0.1mg/ml of bovine serum albumin)]; APB assay [(50mM of sodium phosphate buffer (pH 7.4),150 mM NaCl, puromycin (40µM) and 0.1mg/ml of bovine serum albumin, pH 6.5)]; PSA assay [(50mM of sodium phosphate buffer (pH 7.4), puromycin (40µM) and 0.1mg/ml of bovine serum albumin].
The reaction was initiated by adding 30μL (APB and PSA) or 50μL (PGI) of tissue or plasma sample to 1mL of the assay mixture. This was incubated at 37ºC for 30 minutes and the reaction was stopped by addition of 1 mL of 0.1M sodium acetate buffer (pH 4.2). The excitation and emission wavelengths were 345 and 412 nm, respectively. Blanks were used to determine background fluorescence. Relative fluorescence was converted into picomoles of product using a standard curve constructed with increasing concentrations of β-naphthylamine.
Protein concentration was measured in triplicate by the Bradford method [28], using BSA (1 mg/mL) as calibrator. Results from the CRC tissues and from plasma samples were recorded as units of peptidase per milligram of protein (UP/mg prot) and per liter of plasma (UP/L), respectively. One unit of peptidase activity (UP) is the amount of enzyme required to release one pmol of β-naphthylamine per minute. Fluorogenic assays were linear with respect to hydrolysis time and protein content.
Real-time quantitative PCR analysis
Quantitative RT-PCR for detecting PGPEP1 (PGI), RNPEP (APB) and NPEPPS (PSA) mRNA was performed to determine the transcription levels of these peptidases. The RNA of tumor and nontumor tissue samples from CRC patients was isolated with the RNeasy Protect kit (Qiagen), including a DNase digestion step using an RNase-free DNase kit (Qiagen) to exclude possible contamination by genomic DNA. First-strand cDNA was synthesized from 25 µg of total RNA from each human sample using Moloney murine leukemia virus reverse transcriptase and random hexamers according to the manufacturer's instructions (first-strand cDNA Synthesis Kit, Amersham Biosciences, Essex, UK). The resulting cDNA samples were amplified by PCR (7900 Real-Time PCR System, Applied Biosystems) with specific oligonucleotide primer pairs designed with the analysis software Primer 3 and synthesized by Sigma-Genosys (Cambridge, UK).
Based on previous experiments on CRC [17], human renal cell carcinoma [21] and other human tissues [29], TATA box binding protein (TBP), peptidylprolyl isomerase A (PPIA), Hypoxantine phosphoribosyltransferase 1 (HPRT1), Polymerase(RNA)II(DNA directed) polypeptide A (POLR2 A), Glyceraldehyde-3´´-phosphate dehydrogenase (GAPDH) and succinate dehydrogenase complex subunit A (SDHA) were chosen as endogenous reference genes. The sequences of the primers used to amplify PGI, APB, PSA and the six housekeeping genes are shown in Table 2. All primers were synthesized and purified by Sigma-Genosys (Cambridge, UK).
Sequence of forward (F) and reverse primers (R) of indicated target genes and the size expected for each PCR-amplified product. Primers for the assayed housekeeping genes are also shown.
| Enzyme/HKG | Gene Symbol | Forward Primer | Reverse Primer | Amplicon size (bp) |
|---|---|---|---|---|
| Piroglutamyl aminopeptidase | PGPEP1 | 5´-GCCCTGTGGGAGAAGCAC-3´ | 5´-AGCCCCTTGTAGCCCTTGTT-3´ | 110 |
| Aminopeptidase B | RNPEP | 5´-CGAAGCATCTTAGCCGATGAC-3´ | 5´-TCGATCAAACTCAAAACCTGGA-3´ | 102 |
| Aminopeptidase puromycin sensitive | NPEPPS | 5´-CGTTAGAAGAAGCCCGTCGT-3´ | 5´-AAAGTAGTGCCATCACCATGCTT-3´ | 118 |
| TATA box binding protein | TBP | 5´-GGATAAGAGAGCCACGAACCAC-3´ | 5´-TTAGCTGGAAAACCCAACTTCTG-3´ | 139 |
| Succinate dehydrogenase complex, subunit A | SDHA | 5´-TCTGCCCACACCAGCACT-3´ | 5´-CCTCTCCACGACATCCTTCC-3´ | 142 |
| Peptidylpropyl isomerase A | PPiA | 5´-GGTCCCAAAGACAGCAGAAAA-3´ | 5´-TCACCACCCTGACACATAAACC-3´ | 114 |
| Hypoxantine phosphoribosyltransferase 1 | HPRT1 | 5´-GCCAGACTTTGTTGGATTTGA-3´ | 5´-GGCTTTGTATTTTGCTTTTCC-3´ | 130 |
| Polymerase(RNA)II(DNA directed) polypeptide A | POLR2A | 5´-ACATCACTCGCCTCTTCTACTCC-3´ | 5´-GTCTTGTCTCGGGCATCGT-3´ | 268 |
| Glyceraldehyde-3´´-phosphate dehydrogenase | GAPDH | 5´-CAATGCCTCCTGCACCAC-3´ | 5´-CCTGCTTCACCACCTTCTTG-3´ | 350 |
Statistical analysis
Kolmogorov-Smirnov and Shapiro-Wilk tests were applied to data obtained from tissue and plasma samples respectively to know if the numbers followed or not a normal distribution. Based on this information (p<0.05), peptidase activities in tissue and plasma were analyzed with non parametric probes: Mann-Whitney test and Kruskal-Wallis tests were used to detect differences between two or three groups, respectively. Spearman`s correlation test was performed to evaluate the correlation between tissue and plasmatic activities and with tumor size and patients' gender.
Finally, Kaplan-Meier curves and log-rank test were performed to evaluate the association between soluble peptidase activities and 5-year survival. The compared groups were created by cut-off points based on median enzyme activity values, as previously reported [30,31]. A Cox regression model was used to test the independent effects of clinical and pathological variables and peptidase activity on five-year overall survival. SPSS® 21.0 software was used for the statistical analysis.
Results
Peptidase activity and mRNA expression in tissue specimens
APB and PSA peptidase activity was significantly higher in tumor samples (adenoma and adenocarcinoma) than in the adjacent non-tumor tissue (Kruskal-Wallis, p=0.001 for APB and p=0.0001 for PSA). In the adenomas the activity was near to three and two-fold higher than the uninvolved mucosa, respectively. PGI activity was higher in adenocarcinomas, however it did not reach statistical significance (Kruskal-Wallis, p= 0.2). Results are summarized in Table 3A.
Peptidase activity in tissue from CRC patients. Values are means + SE of peptidase activity recorded as pmol of units of peptidase (UP) per milligram of protein. a: Adenocarcinoma vs Adenoma; b: Adenocarcinoma vs Normal; c: Adenoma vs Normal.
| Peptidase | UP/L (Mean ± S.E) | Mann- Whitney (p =) |
|---|---|---|
| APB | ||
| Adenocarcinoma | 13298 ± 693 | 0.073a |
| Adenoma | 27812 ± 4202 | 0.002b |
| Non-tumoral | 10952 ± 1531 | 0.001c |
| PSA | ||
| Adenocarcinoma | 15355 ± 621 | 0.443a |
| Adenoma | 19747 ± 2296 | 0.0001b |
| Non-tumoral | 10587 ± 880 | 0.001c |
| PGI | ||
| Adenocarcinoma | 176 ± 14.35 | 0.145a |
| Adenoma | 138 ± 12.02 | 0.156b |
| Non-tumoral | 151 ± 18.38 | 0.934c |
These differences were not correlated with mRNA levels. Thus, PGPEP1 (PGI) and NPEPPS (PSA) were downregulated in tumor specimens compared with non-neoplasic tissue (Kruskal-Wallis, p<0.001 for both genes) and RNPEP (APB) mRNA levels were similar throughout the uninvolved mucosa-adenoma-adenocarcinoma sequence (Table 3B).
Peptidase mRNA expression. Values are means ± SE of peptidase mRNA expression recorded as relative units. a: Adenocarcinoma vs Adenoma; b: Adenocarcinoma vs Normal; c: Adenoma vs Normal.
| Peptidase | Relative Units (Mean ± S.E ) | Mann-Whitney (p =) |
|---|---|---|
| RNPEP (APB) | ||
| Adenocarcinoma | 163.93 ± 34.81 | 0.144a |
| Adenoma | 174.8 ± 15.76 | 0.797 b |
| Non-tumoral | 153.76 ± 26.03 | 0.312 c |
| NPEPPS (PSA) | ||
| Adenocarcinoma | 107.89 ± 15.36 | 0.001a |
| Adenoma | 36.06 ± 6.2 | 0.029b |
| Non-tumoral | 166.29 ± 20.2 | 0.0001c |
| PGPEP1 (PGI) | ||
| Adenocarcinoma | 43.15 ± 7.17 | 0.005a |
| Adenoma | 13.73 ± 2.12 | 0.001b |
| Non-tumoral | 157.07 ± 41.39 | 0.0001c |
Peptidase activity in CRC tissue according to clinicopathological variables and topographic distribution
Adenocarcinomas with low grades showed higher PGI activity than high grade tumors (Mann-Whitney, p=0.011). A similar tendency was observed in CRCs showing lymphatic invasion, although this result was not significant (Mann-Whitney, p=0.05). PSA and APB activities did not revealed significant changes. These results are summarized in Table 1A.
Topographic distribution of adenocarcinomas displayed significant differences in all the analyzed peptidases, with higher activities in adenocarcinomas located in the colon than in the rectum (Mann-Whitney, p<0.05 for all cases) (Table 1A). To elucidate if these significant differences between the two topographies, colon and rectum, were related with the chemo-radiotherapy regimes previously administered to some patients with rectal adenocarcinoma, we compared enzyme activities in cases with and without this previous treatment. However, it did not throw any significant result (Mann-Whitney, p > 0.05 in all cases).
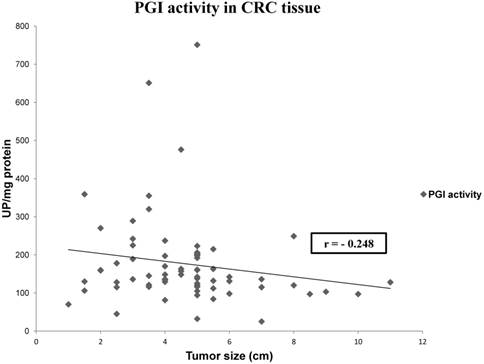
We also analyzed if there is any correlation between tissue and plasmatic activities, and between tumor size and patients' gender and enzyme activities. We only found a statistically significant correlation between tissue PGI activity and tumor size (Spearman Rho test, r= -0.248, p= 0.04) (Figure 1).
Peptidase activity in tissue according to overall survival
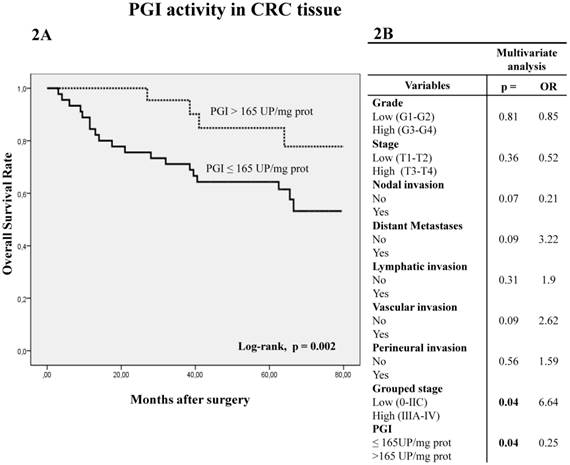
Kaplan-Meier curves and log-rank test showed that five-year overall survival (OS) of patients with CRC was better when PGI activity was higher than 165 UP/mg of protein (log-rank p=0.002) (Figure 2A). A similar trend was observed with APB and PSA (cut-off value 15000 UP/mg for both enzyme activities), when five-year OS was analyzed by log-rank test, although it did not reach statistical significance (p=0.07 and p=0.057, respectively).
Multivariate Cox regression model showed that PGI activity (p=0.04) and grouped stage (p=0.04) were independent factors influencing patient OS (Fig 2B).
Representation of the correlation between PGI activity and tumor size in CRC. Values represent mean ± SE of enzyme activity recorded as units of enzyme per milligram of protein (UP/mg prot). Spearman's rho test: coefficient (r) = -0.248; p< 0.05.
Kaplan-Meier curves and Multivariate Analysis (Cox regression model) with PGI activity in CRC tissues. Overall survival (2A) of 81 CRC patients according to their tumor PGI activity pattern. Multivariate (2B) analysis of clinicopathological variables and tumor PGI activity in predicting overall survival of patients with CRC.
Peptidase activity in plasma samples compared with controls and according to overall survival and other clinicopathological variables
Plasma from CRC patients and healthy individuals showed similar peptidase activities (Mann-Whitney test p>0.05) (Table 4). The stratification of data according to clinicopathological variables also showed no significant changes (Table 1B).
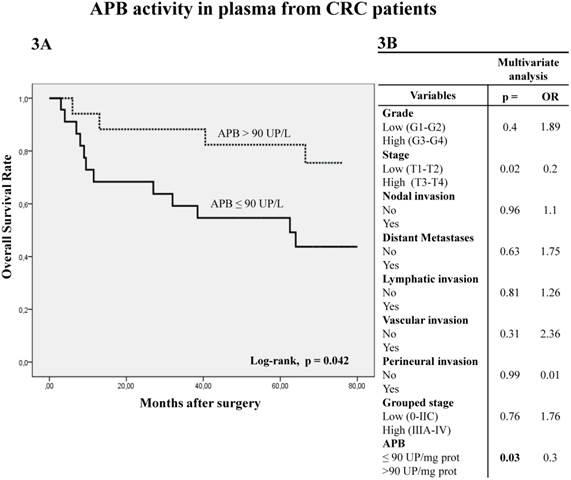
The analysis of APB activity in plasma samples displayed similar results to those obtained in tumor tissue. Thus, when plasmatic APB activity was higher than 90 UP/L the OS was significantly better in Kaplan-Meier curves (Fig 3A) (log-rank p=0.042). Multivariate analysis showed that APB activity (p=0.03) and local invasion (p=0.02) are independent prognostic factors of OS (Fig 3B).
Discussion
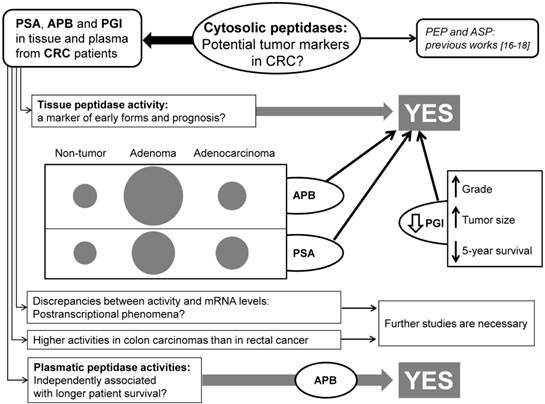
Many studies have revealed that the altered expression and activity of several peptidases may play a role in tumor transformation and progression of CRC. Thus, some authors have proposed that the analysis of these enzymes may have a predictive value in the early detection and prognosis of patients with CRC [10,16,17,31-32]. In this sense, the first studies pointed to cell-surface peptidases such as dipeptidyl-peptidase IV (DPPIV/CD26) [10], aminopeptidase N (APN/CD13) [31] and fibroblast activation protein (FAP) [32] as a new tumor markers and promising targets for CRC therapy. However, we recently reported that cytosolic peptidases may be also of interest in the searching of new markers and targets for this disease [16,17]. So, to throw more light to this question, in this study we analyzed the activity and mRNA expression of three cytosolic enzymes (APB, PSA and PGI) in colorectal tissues and plasma from a series of CRC patients (see the flow-chart in the Figure 4).
Peptidase activity in plasma from CRC patients. Values are means ± SE of units of peptidase per liter of plasma (UP/L).
| Peptidase | UP/L (Mean ± S.E ) | Mann-Whitney (p =) |
|---|---|---|
| APB | ||
| Patient | 95.37 ± 5.34 | 0.106 |
| Control | 105.29 ± 6.57 | |
| PSA | ||
| Patient | 338.07 ± 30.14 | 0.873 |
| Control | 340.71 ± 48.47 | |
| PGI | ||
| Patient | 21.63 ± 3.11 | 0.988 |
| Control | 18.61 ± 3.29 |
Kaplan-Meier curves and Multivariate Analysis (Cox regression model) with APB activity in plasma from CRC patients. Overall survival (3A) of 40 CRC patients according to their plasmatic APB activity pattern. Multivariate (3B) analysis of clinicopathological variables and tumor APB plasmatic activity in predicting overall survival of patients with CRC.
Flow-chart summarizing the most important findings of this study.
The adenoma-adenocarcinoma sequence in the large bowel describes that the gradual progression from normal to dysplastic epithelium, and hence to carcinoma, is associated with the accumulation of genetic imbalances [5,6]. It has long been reported that expression of several proteases, such as matrix metalloproteinases (MMPs) and peptidases, increases gradually throughout the sequence, playing an important role in the development and progression of these tumors [33,34]. Furthermore, some studies have suggested that the coordinated action of MMPs and aminopeptidases is important for the progression of several neoplastic diseases [8,35]. In a previous work, we observed that PEP activity increased significantly in the preneoplastic adenomatous lesions, then stabilizing in adenocarcinomas [18]. In the present study, 16 adenoma-adenocarcinoma sequences from the same patients were analyzed. Spectrofluorimetric assay showed that PSA and APB activity was increased significantly in colorectal tumors, but particularly in the adenomas, where it was two and three-fold higher (respectively) than in the adjacent uninvolved mucosa.
On the other hand, the analysis of CRC tissues also showed that the activity of PGI correlated negatively with the histological grade and tumor size. Moreover, 5-year overall survival of CRC patients was significantly better when tissue PGI activity was higher. In our previous analysis with ASP and PEP, higher tissue activities also correlated with less aggressive forms and with better patient survival [16,17]. All this findings taken together suggest that the tissue activity of these cytosolic enzymes may be a marker of early forms and prognosis of CRC.
Quantitative RT-PCR assays showed that the mRNA levels were not correlated with the enzyme activity in colorectal tissues. Thus, (PSA) and (PGI) levels were higher in the not cancerous mucosa than in CRC and the adenomatous tissues, and mRNA levels of RNPEP (APB) did not change significantly throughout the adenoma-carcinoma pathway. This discrepancy between mRNA and the protein expression/activity has been described in several studies, which could be due to alterations in mRNA translation efficiency, posttranslational modifications like phosphorylation or acethylation, and mRNA and protein synthesis and degradation rates [8,22,36-38]. Besides, this lack of correlation is widely spread in renal diseases involving altered energy metabolism like diabetes-induced renal hypertrophy, diabetes-induced matrix synthesis or angiotensin II induced renal injury, and hence, it could be a strong possibility for cancer cells to show this discrepancy [38-41]. These phenomena illustrate the importance of not relying only on mRNA level the evaluation of any protein change [38].
Topographic distribution of adenocarcinomas also displayed significant differences in all the analyzed peptidases. Thus, rectal carcinomas showed decreased enzyme activities compared with colonic adenocarcinomas. Recently, we demonstrated a similar distribution of PEP activity in CRC tissues and observed that it was decreased significantly in rectal cancer tissues from patients who received chemo-radiotherapy preoperatively for a better control of the disease [17]. Similar patterns were observed with other proteases, which indicate that these enzymes could be of potential use in clinical practice to predict the response to preoperative chemo-radiotherapy [42,43]. We did not observe any change in the activity of PSA, APB and PGI in rectal cancer in response to preoperative treatment and, therefore, further studies are necessary to know the cause of these topographic differences.
Previous studies in a wide variety of tumors have demonstrated that cytosolic peptidases may change their activities depending on the tumor type or on the phase of development where the tumor is. For example, it has been reported increased APB, PSA and PGI activities in breast [19,20] and thyroid tumors [23] with respect to uninvolved tissues. PSA and PGI activities were also increased in high grade clear cell renal cell carcinomas [22] and higher APB activity was correlated with better prognosis of these patients [11]. However, these enzymes did not change their activity in head and neck cancers [41]. So, these data support the idea that tumor growth regulation by peptidases occur in a tumor specific manner [8].
Cytosolic and cell-surface peptidases can be released to diverse body fluids, including plasma, urine, pleural and peritoneal effusions. Although the origin of these circulating enzymes is still controversial, the comparative studies of peptidase profiles among cancer patients and control subjects have yielded significant results in several studies [10,17,20,30,45-47]. Furthermore, the correlation among plasma/serum values and classic histopathological parameters for tumor aggressiveness and survival have pointed to these enzymes as useful biomarkers for helping in screening, diagnosis, staging, prognosis and monitoring cancer therapy [10,17-20,30,45-47].
In this work we did not found any significant difference in the analyzed peptidases in plasma between CRC patients and healthy individuals. However, higher plasmatic APB activity was independently associated with longer patient survival. Although this result was opposite of that described previously with ASP and PEP, which plasmatic activities correlated positively with poorer survival rate [16,17], these evidences also point to the determination of cytosolic peptidases in plasma as a valuable method in the determination of CRC prognosis. Further studies focusing on the molecular translation of this descriptive analysis with higher number of patients would be advisable to confirm the prognostic significance of these data.
Conclusion
The present study shows for the first time that the activity and mRNA levels of PSA, APB and PGI are altered throughout the colorectal adenoma-carcinoma sequence and that the activity of tissue PGI and plasmatic APB are independent prognostic factors for the 5-year survival of CRC patients. Taken into account that CRC is a major health problem in developed countries, the arrival of cell-surface but also cytosolic peptidases as new diagnostic tools with prognostic impact may be particularly profitable in hospital practice.
Acknowledgements
We wish to thank Arantza Pérez (UPV/EHU) for her technical contribution to this study and to Professor Juan Bilbao (UPV/EHU) for his statistical support. This work was supported by a grant from the Basque Government (IT8-11/13 and S-PE12UN042), the University of the Basque Country UPV/EHU (UFI 11/44) and the Gangoiti-Barrera Foundation.
Competing Interests
The authors declare that there are no competing interests.
References
1. Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J. et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374-1403
2. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-117
3. Zavoral M, Suchanek S, Majek O. et al. Colorectal cancer screening: 20 years of development and recent progress. World J Gastroenterol. 2014;20:3825-3834
4. Raskov H, Pommergaard HC, Burcharth J. et al. Colorectal carcinogenesis-update and perspectives. World J Gastroenterol. 2014;20:18151-18164
5. Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113-130
6. Kim EC, Lance P. Colorectal polyps and their relationship to cancer. Gastroenterol Clin North Am. 1997;26:1-17
7. Heslin MJ, Yan J, Johnson MR. et al. Role of matrix metalloproteinases in colorectal carcinogenesis. Ann Surg. 2001;233:786-792
8. Carl-McGrath S, Lendeckel U, Ebert M. et al. Ectopeptidases in tumor biology: A review. Histol Histopathol. 2006;21:1339-1353
9. Wickström M, Larsson R, Nygren P. et al. Aminopeptidase N (CD13) as a target for cancer chemotherapy. Cancer Sci. 2011;102:501-508
10. Cordero OJ, Ayude D, Nogueira M. et al. Preoperative serum CD26 levels: diagnostic efficiency and predictive value for colorectal cancer. Br J Cancer. 2000;80:1139-1146
11. Larrinaga G, Blanco L, Sanz B. et al. The impact of peptidase activity in clear cell renal cell carcinoma survival. Am J Physiol Renal Physiol. 2012;303:F1584-1591
12. Antczak C, De Meester I, Bauvois B. Ectopeptidases in pathophysiology. Bioessays. 2001;23:251-260
13. Nanus DM, Bogenrieder T, Papandreou CN. et al. Aminopeptidase A expression and enzymatic activity in primary human renal cancers. Int J Oncol. 1998;13:261-267
14. Thompson MW, Hersh LB. The puromycin-sensitive aminopeptidase. Role in neurological, reproductive, inmunological and proliferative disorders. In: (ed.) Hooper NM, Lendeckel U. Aminopeptidases in Biology and Disease. NY: Kluwer Academic/Plenum Press. 2004:1-15
15. Myöhänen TT, Pyykkö E, Männistö PT. et al. Distribution of prolyl oligopeptidase in human peripheral tissues and in ovarian and colorectal s. J Histochem Cytochem. 2012;60:706-715
16. Larrinaga G, Perez I, Ariz U. et al. Clinical impact of aspartyl aminopeptidase expression and activity in colorectal cancer. Transl Res. 2013;162:297-308
17. Larrinaga G, Blanco L, Perez I. et al. Prolyl endopeptidase activity is correlated with colorectal cancer prognosis. Int J Med Sci. 2014;11:199-208
18. Larrinaga G, Perez I, Blanco L. et al. Increased prolyl endopeptidase activity in human neoplasia. Regul Pept. 2010;163:102-106
19. Martínez JM, Prieto I, Ramírez MJ. et al. Aminopeptidase activities in breast cancer tissue. Clin Chem. 1999;45:1797-1802
20. Martínez-Martos JM, del Pilar Carrera-González M, Dueñas B. et al. Renin angiotensin system-regulating aminopeptidase activities in serum of pre- and postmenopausal women with breast cancer. Breast. 2011;20:444-447
21. Varona A, Blanco L, López JI. et al. Altered levels of acid, basic, and neutral peptidase activity and expression in human clear cell renal cell carcinoma. Am J Physiol Renal Physiol. 2007;292:780-788
22. Blanco L, Larrinaga G, Perez I. et al. Acid, basic and neutral peptidases present different profiles in chromophobe renal cell carcinoma and in oncocytoma. Am J Physiol-Renal Physiol. 2008;294:850-858
23. Larrinaga G, Blanco L, Errarte P. et al. Altered peptidase activities in thyroid neoplasia and hyperplasia. Dis Markers. 2013;35(6):825-832
24. Edge SB, Byrd DR, Compton CC. et al. AJCC Cancer Staging Manual, 7th Edition, Springer, New York. 2010.
25. Washington MK, Berlin J, Branton P. et al. Protocol for the examination of specimens from patients with primary carcinoma of the colon and rectum. Arch Pathol Lab Med. 2009;133:1539-1551
26. Olivo RA, Pereira Teixeira PT, Silveira PF. Representative aminopeptidases and prolyl endopeptidase from murine macrophages: comparative activity levels in resident and elicited cells. Biochem Pharmacol. 2005;69:1441-1450
27. Zambotti-Villela L, Yamasaki SC, Villarroel JS. et al. Aspartyl, arginyl and alanyl aminopeptidase activities in the hippocampus and hypothalamus of streptozotocin-induced diabetic rats. Brain Res. 2007;1170:112-118
28. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-254
29. van Hensbergen Y, Broxterman HJ, Hanemaaijer R. et al. Soluble aminopeptidase N/CD13 in malignant and nonmalignant effusions and intratumoral fluid. Clin Cancer Res. 2002;8:3747-3754
30. Javidroozi M, Zucker S, Chen WT. Plasma seprase and DPP4 levels as markers of disease and prognosis in cancer. Dis Markers. 2012;32:309-320
31. Hashida H, Takabayashi A, Kanai M. et al. Aminopeptidase N is involved in cell motility and angiogenesis: its clinical significance in human colon cancer. Gastroenterology. 2002;122:376-386
32. Henry LR, Lee HO, Lee JS. et al. Clinical implications of fibroblast activation protein in patients with colon cancer. Clin Cancer Res. 2007;13:1736-1741
33. Talieri M, Mathioudaki K, Prezas P. et al. Clinical significance of kallikrein-related peptidase 7 (KLK7) in colorectal cancer. Thromb Haemost. 2009;101:741-747
34. Heslin MJ, Yan J, Johnson MR. et al. Role of matrix metalloproteinases in colorectal carcinogenesis. Ann Surg. 2001;233:786-792
35. Petrovic N, Schacke W, Shapiro LH. CD13/Aminopeptidase N in tumor growth and angiogenesis. In: (ed.) Hooper NM, Lendeckel U. Aminopeptidases in Biology and Disease. New York: Kluwer Academic/Plenum Press. 2004:179-200
36. Quaroni A, Nichols BL, Quaroni E. et al. Expression and different polarity of aminopeptidase N in normal human colonic mucosa and colonic tumors. Int J Cancer. 1992;51:404-411
37. Mathieu MG, Linley AJ, Reeder SP. et al. HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun. 2010;10:2-9
38. Kasinath BS, Mariappan MM, Sataranatarajan K. et al. mRNA traslation: unexplored territory in renal science. J Am Soc Nephrol. 2006;17:3281-3292
39. Scacheri PC, Rozenblatt-Rosen O, Caplen N. et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1892-1897
40. Murakami T, Nojiri M, Nakayama H. et al. Post-translational modification is essential for catalytic activity of nitrile hydratase. Protein Sci. 2000;9:1024-1030
41. Mamane Y, Petroulakis E, LeBacquer O. et al. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416-6422
42. Unsal Kilic D, Uner A, Akyurek N. et al. Matrix metalloproteinase-9 expression correlated with response in patients with locally advanced rectal cancer undergoing preoperative chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2007;67:196-203
43. Nishioka M, Shimada M, Kurita N. et al. Gene expression profile can predict pathological response to preoperative chemoradiotherapy in rectal cancer. Cancer Genomics Proteomics. 2011;8:87-92
44. Perez I, Varona A, Blanco L. et al. Increased APN/CD13 and acid aminopeptidase activities in head and neck squamous cell carcinoma. Head & Neck. 2009;31:1335-1340
45. Murakami H, Yokoyama A, Kondo K. et al. Circulating aminopeptidase N/CD13 is an independent prognostic factor in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:8674-8679
46. Goossens F, De Meester I, Vanhoof G. et al. Distribution of prolyl oligopeptidase in human peripheral tissues and body fluids. Eur J Clin Chem Clin Biochem. 1996;34:17-22
47. Cordero OJ, Imbernon M, Chiara LD. et al. Potential of soluble CD26 as a serum marker for colorectal cancer detection. World J Clin Oncol. 2011;2:245-261
Author contact
![]() Corresponding author: Gorka Larrinaga, MD, PhD, Department of Nursing I, University School of Nursing, University of the Basque Country. P.O. Box 699, 48080, Bilbao, Bizkaia, Spain, Fax: +34 94 601 3580, Phone: +34 94 601 8076, email: gorka.larrinagaeus
Corresponding author: Gorka Larrinaga, MD, PhD, Department of Nursing I, University School of Nursing, University of the Basque Country. P.O. Box 699, 48080, Bilbao, Bizkaia, Spain, Fax: +34 94 601 3580, Phone: +34 94 601 8076, email: gorka.larrinagaeus

 Global reach, higher impact
Global reach, higher impact