Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(2):126-134. doi:10.7150/ijms.9951 This issue Cite
Research Paper
Single Nucleotide Polymorphism in Ag85 Genes of Mycobacterium Tuberculosis Complex: Analysis of 178 Clinical Isolates from China and 13 BCG strains
1. State Key Laboratory for Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, P. R. China
2. Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou 310003, China
3. Immunology Department, Changzhi Medical College, Shanxi, China
4. Beijing Center for Diseases Prevention and Control, Beijing 100013, China
* Yi Jiang, Haican Liu and Machao Li contributed equally to this study.
Received 2014-6-20; Accepted 2014-12-1; Published 2015-1-5
Abstract
Host immune pressure and associated immune evasion of pathogenic bacteria are key features of host-pathogen co-evolution. Human T-cell epitopes of Mycobacterium tuberculosis (M. tuberculosis) were evolutionarily hyperconserved and thus it was deduced that M. tuberculosis lacks antigenic variation and immune evasion. However, in our previous studies, proteins MPT64, PstS1, Rv0309 and Rv2945c all harbored higher numbers of amino acid substitutions in their T cell epitopes, which suggests their roles in ongoing immune evasion. Here, we used the same set of 180 clinical M. tuberculosis complex (MTBC) isolates from China, amplified the genes encoding Ag85 complex, and compared the sequences. The results showed that Ag85 were hyperconserved in T/B cell epitopes and the genes were more likely to be under purifying selection. The divergence of host immune selection on different proteins may result from different function of the proteins. In addition, A312G of Ag85A and T418C of Ag85B may represent special mutations in BCG strains, which may be used to differentiate M.bovis and BCG strains from MTB strains. Also, C714A in Ag85B seems to be a valuable phylogenetic marker for Beijing strains.
Keywords: Genetic diversity, Mycobacterium tuberculosis, Ag85
INTRODUCTION
Tuberculosis (TB) is one of the most important issues of public health worldwide. About one third of the world population has been infected with M. tuberculosis, over 8.7 million new cases and 1.4 million deaths each year (1). The current efforts to reduce the global problem have been focused on improving the diagnosis methods and effective vaccines. The biochemical, immunological, and molecular biological characteristics of M. tuberculosis have led to the identification of several antigens which may be useful in the development of improved diagnostic methods and/or vaccines (2).
In 2010, Inaki Comas et al reported that human T cell epitopes of M. tuberculosis were evolutionarily hyperconserved and thus deduced that M. tuberculosis was lack of antigenic variation and immune evasion (3). However, our previous studies showed that there were polymorphisms existing in two important antigens, MPT64 (4) and PstS1 (5) in clinical M. tuberculosis strains isolated from China. This may be the reason for changes in the antigens produced, which may in turn cause alteration of related functions, thereby allowing immune evasion. Some other proteins such as Rv2945c and Rv0309 also owned polymorphisms, which suggest their roles in diversifying selection to evade host immunity (6). The antigen 85 complex (Ag85) consists of three predominantly secreted proteins (Ag85A, Ag85B, and Ag85C), which plays a key role in the mycobacterial pathogenesis and also possesses enzymatic mycolyltransferase activity involved in cell wall synthesis (7). Disruption of the gene encoding Ag85A in M. tuberculosis produces a strain that fails to replicate in human or mouse macrophages indicating that Ag85A may play a key role in M.tuberculosis pathogenesis. Knockout of the gene encoding Ag85C results in 40% reduction of M.tuberculosis the cell wall mycoloylation. Ag85 complex contribute to adherence, invasion, and dissemination of mycobacteria in host cells (8). By virtue of their strong potential to induce Th1-type immune responses, important for the control of intracellular infections, Mycobacterium Ag85 complex rank among the most promising TB vaccine candidate antigens. (9-14). Recently, Modified-Vaccinia-Ankara (MVA)85A vaccine became the first TB vaccine since BCG itself to complete an efficacy trial (15).
Here, we used the same set of clinical M. tuberculosis complex (MTBC) isolates(including two BCG strains) from China in our previous study (4), amplified genes of the antigens Ag85 (Ag85A, Ag85B, and Ag85C) and compared the sequences to explore the genetic diversity of them and to evaluate the impact of immune recognition on sequence variation of these three genes. In addition, we analyzed changes in protein level which was induced by single nucleotide polymorphism in Ag85 genes.
MATERIALS AND METHODS
Strains and DNA preparation
The first set of strains consisted of 180 clinical isolates that were selected from 2346 MTBC strains isolated in China genotyped by spoligotyping previously (16). All major and rare genotyping strains in China were included (Table 1). Considering the predominance of the Beijing family strains in China, we chose about half of the Beijing family strains (92 strains) and half non-Beijing family strains (88 strains). We randomly selected the 92 Beijing family strains from 1738 Beijing strains among 2346 strains. The other 88 strains were selected from 608 non-Beijing family isolates. Further, we attempted to purposely include strains representing different spoligotypes that were isolated from different regions. Table 2 showed the numbers of strains used in this study that were obtained from different provinces in China. A second set of strains contained 11 BCG strains, each of which originated from different places around the world. The strain names were showed in Table 3.
No. of the strains of each Spoligotype pattern
| Spoligotyping | No. of strains |
|---|---|
| Beijing | 92 |
| T | 13 |
| U | 28 |
| MANU | 11 |
| Haarlem | 5 |
| EAI | 1 |
| LAM | 2 |
| H37Rv family | 1 |
| BCG | 2 |
| S | 1 |
| CAS | 4 |
| new | 20 |
No. of the strains of different provinces in China
| Places | No. of isolates |
|---|---|
| Anhui Province | 12 |
| Shannxi Province | 17 |
| Beijing Municipality | 11 |
| Fujian Province | 29 |
| Gansu Province | 12 |
| Guangxi Zhuang Autonomous Region | 29 |
| Sichuan Province | 1 |
| Henan Province | 12 |
| Hunan Province | 7 |
| Xizang (Tibet) Autonomous Region, | 11 |
| Xinjiang Uygur Autonomous Region | 13 |
| Jilin Province | 14 |
| Zhejiang Province | 12 |
Strains of Mycobacterium bovis and Bacillus Calmette Guerin (BCG).
| ID No. | Strain name |
|---|---|
| 1 | BCG Birkhaug |
| 2 | BCG China |
| 3 | BCG Danish |
| 4 | BCG Frappier |
| 5 | BCG Glaxo |
| 6 | BCG Moreau |
| 7 | BCG Phipps |
| 8 | BCG Prague |
| 9 | BCG Swedens |
| 10 | BCG Tice |
| 11 | BCG Russia |
| 12 | BCG Tokyo* |
| 13 | BCG Paster* |
| 14 | BCG Mexco* |
| 15 | M. bovis AF2122/97* |
* Data were obtained from the NCBI genome website
These strains were cultured using the standard Löwenstein-Jensen medium method, the genomic DNA were prepared according to previously reported and then used directly in polymerase chain reactions (PCRs).
The following Ag85 genes of the four published M.bovis and BCG strains were obtained from the NCBI genome website: M. bovis AF2122/97 (NC_002945), BCG Pasteur 1173P2 (NC_008769), BCG Tokyo 172 (NC_012207) and BCG Mexico (NC_016804).
Primers
The nucleotide sequences of the primers (from the 5' to 3' end) used in this study were designed with DNAstar software according to H37Rv genome sequence and showed in Table 4.
The primers used in this study for PCR amplification
| Gene | Locus tag | Length(bp) | Primers |
|---|---|---|---|
| Ag85A | Rv3804c | 1166 | 5'- CACCGCCGCTAGATGTTGTG-3'F |
| 5'- CGCCCGAAGTTGTGGTTGAC-3'R | |||
| Ag85B | Rv1886c | 1234 | 5'- ACTCGGCTAACTGGCTGGT-3'F |
| 5'- CGGTAACCGATACGGAAATG -3'R | |||
| Ag85C | Rv0129c | 1509 | 5'- TGGTCGGCAGTAAGCATAGG-3'F |
| 5'- ACTGGTTGGGAGCGGCC -3'R |
Polymerase Chain Reaction
The PCR were performed in a total volume of 20μl. The PCR mix contained 10μl PCR buffer, 100nM each primer, 200μM each of the four dNTPs and 0.5U DNA Taq Polymerase (Takara). An initial denaturation of 5min at 94℃ was followed by 35 cycles of denaturation at 94℃ for 45s, annealing at 62℃ for 45s and extension at 72℃ for 1min, followed by a final extension at 72℃ for 10min.
Negative controls using ddH2O instead of DNA were included each time when the PCR was performed. The positive control was 500pg DNA from M. tuberculosis H37Rv. The presence and size of each PCR product were determined by electrophoresis on 2% agarose gel in Tris/boric acid/EDTA buffer followed by staining with ethidium bromide.
We performed all of the PCRs at least twice to validate the reproducibility. The variants were confirmed by sequencing of the new PCR products.
Sequence and data Analysis
The sequences of the PCR products were determined by ABI 3730xl DNA Analyzer.
The sequences were first aligned by ClustalW (17) software with the Ag85 genes sequence from M. tuberculosis H37Rv genome to determine the regions of the genes, and then these regions were split out by a personalized PERL script. The sequence compare and translation were carried out by Bioedit software. Values of dN and dS were calculated by MEGA5. In addition, SPSS 14.0 (SPSS, Inc.) was used to perform chi-square analysis, and differences were considered to be statistically significant when P<0.05.
RESULTS
Mutations in gene sequences of Ag85
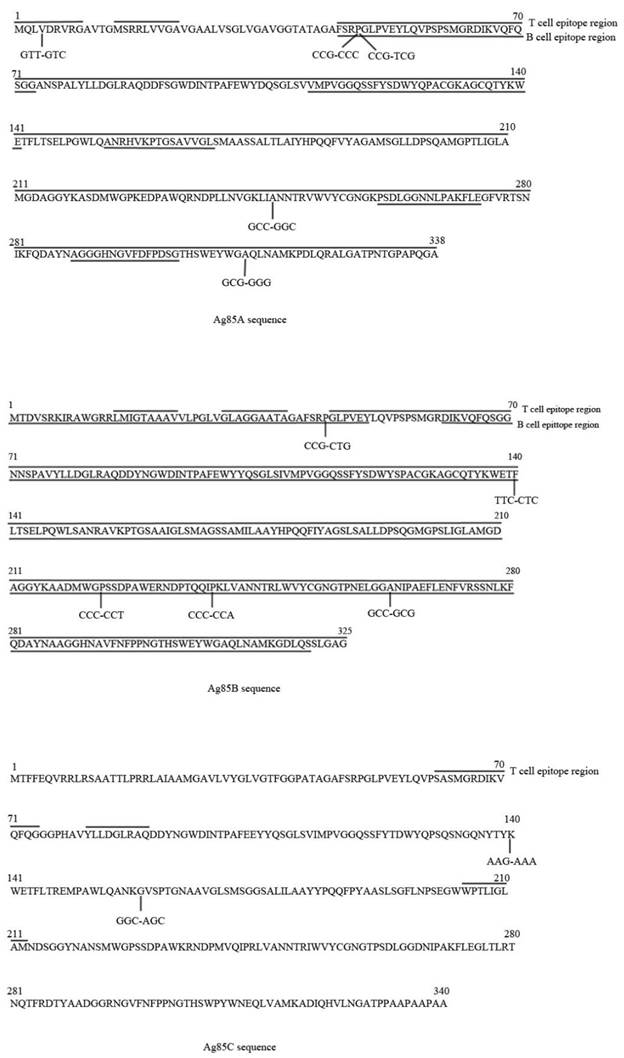
All 180 strains presented relative PCR products of antigens Ag85A, Ag85B and Ag85C. Table 5 showed the mutations in the gene sequences of Ag85A, Ag85B and Ag85C. Ag85A harbored three nonsynonymous mutations and two synonymous mutations. Ag85B owned two nonsynonymous mutations and three synonymous mutations. There were one nonsynonymous mutation and one synonymous mutation in antigen Ag85C.
A total of 16 BCG strains (14 different BCG strains described above and 2 BCG strains from the clinical sample in China in the first isolates set) and one M.bovis (M. bovis AF2122/97, NC_002945) were included in this study. All of the M.bovis and BCG strains had two unique SNPs located in C935G of Ag85A and T418C of Ag85B. In the gene sequence of Ag85B, position 714 presented higher polymorphisms, as 93 strains owned an sSNPs (C-A). Six strains, i.e. four CAS family strains, one Beijing strain and one New strain presented same nonsynonymous mutation (G472A) in gene sequence of antigen Ag85C.
Changes in antigen Ag85A, Ag85B and Ag85C among 180 clinical strains*
| Genes | Isolates | Base change | AA change | Spoligotypes |
|---|---|---|---|---|
| Ag85A | FJ05009 | T12C | No change | New |
| FJ06038 ShanX05098 | C139T G141C | P47S No change | Haarlem Beijing | |
| HuN06009 | C734G | A245G | Beijing | |
| FJ07113 | C935G | A312G | BCG | |
| JL06005 | ||||
| Ag85B | AH03031 | C131T | P44L | Beijing |
| FJ07113 | T418C | F140L | BCG | |
| JL06005 | ||||
| GS05127 | C666T | No change | Beijing | |
| GS05129 | ||||
| 93 strains | C714A | No change | -# | |
| AH03037 | C786G | No change | Beijing | |
| Ag85C | JL06007 | G420A | No change | Beijing |
| FJ05009 | G472A | G158S | New | |
| FJ06159 | Beijing | |||
| XZ06003 | CAS | |||
| XJ06018 | CAS | |||
| XJ06153 | CAS | |||
| XJ06188 | CAS |
*: Use the CDS of Ag85A, Ag85B and Ag85C of M. tuberculosis H37Rv strain as the reference sequence.
#: Details are showed in Supplementary Material: Table S1
Changes in T/B cell epitopes
Table 6 presented T cell epitopes and B cell epitopes in Ag85A, Ag85B and Ag85C according to the Immune Epitopes Database (IEDB) (18). In the IEDB database, the impact on immune recognition was tested by bioinformatic approaches, and not in vivo or in vitro. Ag85A and Ag85B owned both T cell epitopes and B cell epitopes, while Ag85C only had three T cell epitopes (See Figure 1). For Ag85A and Ag85B, some epitopes were both T cell epitopes and B cell epitopes. Among all of the strains in this study, 12 of 64 T cell epitopes accounting for 18.75% and one of seven B cell epitopes accounting for 14.29% in Ag85A showed AA changes resulting from nucleotide alterations. Ag85B owned 37 B cell epitopes and 54 T cell epitopes, while 23 of them are both. Three B cell epitopes and three T cell epitopes altered from AA changes in Ag85B. There was no change in T cell epitopes of Ag85C.
Amino acid changes of human T/B cell epitopes in antigen Ag85A, Ag85B and Ag85C*,‡
| T or B epitope | IEDB_ID | Epitope | Rv locus | Base change | AA change | Antigen |
|---|---|---|---|---|---|---|
| T | 56994 | SASMGRDIKVQFQG | Rv0129c | No | No | Ag85C |
| T | 72965 | WPTLIGLAM | Rv0129c | No | No | Ag85C |
| T | 74768 | YLLDGLRAQ | Rv0129c | No | No | Ag85C |
| B | 503 | AAVVLPGLVGLAGGAATAGA | Rv1886c | No | No | Ag85B |
| B | 34776 | LAGGAATAGAFSRPGLPVEY | Rv1886c | CCG-TGG | P-W | Ag85B |
| B | 42790 | MTDVSRKIRAWGRRLMIGTA | Rv1886c | No | No | Ag85B |
| B | 43514 | NDPTQQIPKLVANNTRLWVY | Rv1886c | CCC-CCA | No | Ag85B |
| B | 48646 | PNGTHSWEYWGAQ | Rv1886c | No | No | Ag85B |
| B | 72515 | WGRRLMIGTAAAVVLPGLVG | Rv1886c | No | No | Ag85B |
| B | 103272 | IGLSMAGSSAMILAA | Rv1886c | No | No | Ag85B |
| B | 103457 | PAEFLENFVRSSNLK | Rv1886c | No | No | Ag85B |
| B | 103530 | QSGGNNSPAVYLLDG | Rv1886c | No | No | Ag85B |
| B | 103532 | QSSFYSDWYSPACGK | Rv1886c | No | No | Ag85B |
| B | 103578 | SAAIGLSMAGSSAMI | Rv1886c | No | No | Ag85B |
| B | 103668 | TSELPQWLSANRAVK | Rv1886c | No | No | Ag85B |
| B | 103729 | WGPSSDPAWERNDPT | Rv1886c | CCC-CCT | No | Ag85B |
| B | 103732 | WLSANRAVKPTGSAA | Rv1886c | No | No | Ag85B |
| T | 3094 | AMGDAGGYK | Rv1886c | No | No | Ag85B |
| T | 5623 | AVYLLDGLR | Rv1886c | No | No | Ag85B |
| T | 8685 | DIKVQFQSG | Rv1886c | No | No | Ag85B |
| T | 13215 | ELPQWLSANR | Rv1886c | No | No | Ag85B |
| T | 13473 | ENFVRSSNL | Rv1886c | No | No | Ag85B |
| T | 15116 | EYWGAQLNAMKGDLQSSLGA | Rv1886c | No | No | Ag85B |
| T | 16332 | FIYAGSLSA | Rv1886c | No | No | Ag85B |
| T | 16333 | FIYAGSLSAL | Rv1886c | No | No | Ag85B |
| T | 16924 | FLTSELPQW | Rv1886c | TTC-CTC | F-L | Ag85B |
| T | 18276 | FVRSSNLKF | Rv1886c | No | No | Ag85B |
| T | 21078 | GLPVEYLQV | Rv1886c | No | No | Ag85B |
| T | 21275 | GMGPSLIGL | Rv1886c | No | No | Ag85B |
| T | 21780 | GPSLIGLAM | Rv1886c | No | No | Ag85B |
| T | 26269 | IGLSMAGSSAMILAAY | Rv1886c | No | No | Ag85B |
| T | 27786 | IPAEFLENF | Rv1886c | No | No | Ag85B |
| T | 27901 | IPKLVANNT | Rv1886c | CCC-CCA | No | Ag85B |
| T | 29558 | IYAGSLSAL | Rv1886c | No | No | Ag85B |
| T | 32213 | KLVANNTRL | Rv1886c | No | No | Ag85B |
| T | 38049 | LMIGTAAAV | Rv1886c | No | No | Ag85B |
| T | 42342 | MPVGGQSSF | Rv1886c | No | No | Ag85B |
| T | 43950 | NFVRSSNLKFQDAYNAAGGH | Rv1886c | No | No | Ag85B |
| T | 49862 | PVEYLQVPSPSMGRD | Rv1886c | No | No | Ag85B |
| T | 52025 | QQFIYAGSLSALLDPSQGM | Rv1886c | No | No | Ag85B |
| T | 59627 | SMAGSSAMI | Rv1886c | No | No | Ag85B |
| T | 60262 | SPSMGRDIKVQFQS | Rv1886c | No | No | Ag85B |
| T | 67695 | VANNTRLWVYCGNGT | Rv1886c | No | No | Ag85B |
| T | 73306 | WYYQSGLSI | Rv1886c | No | No | Ag85B |
| T | 76455 | YWGAQLNAMKGDLQSSLGAG | Rv1886c | No | No | Ag85B |
| T | 92817 | GLAGGAATA | Rv1886c | No | No | Ag85B |
| T | 174019 | VEYLQVPSPSMGRDI | Rv1886c | No | No | Ag85B |
| T | 174021 | VPSPSMGRDIKVQFQ | Rv1886c | No | No | Ag85B |
| T/B | 223 | AAIGLSMAGSSAMILAAYHP | Rv1886c | No | No | Ag85B |
| T/B | 1545 | AGGYKAADMWGPSSDPAWER | Rv1886c | CCC-CCT | No | Ag85B |
| T/B | 2695 | ALLDPSQGMGPSLIGLAMGD | Rv1886c | No | No | Ag85B |
| T/B | 3400 | ANRAVKPTGSAAIGLSMAGS | Rv1886c | No | No | Ag85B |
| T/B | 6323 | CGNGTPNELGGANIPAEFLE | Rv1886c | No | No | Ag85B |
| T/B | 8688 | DIKVQFQSGGNNSPAVYLLD | Rv1886c | No | No | Ag85B |
| T/B | 10841 | DWYSPACGKAGCQTYKWETF | Rv1886c | TTC-CTC | F-L | Ag85B |
| T/B | 18700 | GANIPAEFLENFVRSSNLKF | Rv1886c | GCC-GCG | No | Ag85B |
| T/B | 18898 | GCQTYKWETFLTSELPQWLS | Rv1886c | TTC-CTC | F-L | Ag85B |
| T/B | 21096 | GLRAQDDYNGWDINTPAFEW | Rv1886c | No | No | Ag85B |
| T/B | 21797 | GPSSDPAWERNDPTQQIPKL | Rv1886c | CCC-CCT | No | Ag85B |
| T/B | 40165 | LTSELPQWLSANRAVKPTGS | Rv1886c | No | No | Ag85B |
| T/B | 43332 | NAVFNFPPNGTHSWEYWGAQ | Rv1886c | No | No | Ag85B |
| T/B | 45250 | NNSPAVYLLDGLRAQDDYNG | Rv1886c | No | No | Ag85B |
| T/B | 49421 | PSLIGLAMGDAGGYKAADMW | Rv1886c | No | No | Ag85B |
| T/B | 49872 | PVGGQSSFYSDWYSPACGKA | Rv1886c | No | No | Ag85B |
| T/B | 50442 | QDAYNAAGGHNAVFNFPPNG | Rv1886c | No | No | Ag85B |
| T/B | 52026 | QQFIYAGSLSALLDPSQGMG | Rv1886c | No | No | Ag85B |
| T/B | 56895 | SAMILAAYHPQQFIYAGSLS | Rv1886c | No | No | Ag85B |
| T/B | 64079 | THSWEYWGAQLNAMKGDLQS | Rv1886c | No | No | Ag85B |
| T/B | 67697 | VANNTRLWVYCGNGTPNELG | Rv1886c | No | No | Ag85B |
| T/B | 72314 | WDINTPAFEWYYQSGLSIVM | Rv1886c | No | No | Ag85B |
| T/B | 76584 | YYQSGLSIVMPVGGQSSFYS | Rv1886c | No | No | Ag85B |
| B | 1522 | AGGGHNGVFDFPDSG | Rv3804c | No | No | Ag85A |
| B | 3402 | ANRHVKPTGSAVVGL | Rv3804c | No | No | Ag85A |
| B | 49333 | PSDLGGNNLPAKFLE | Rv3804c | No | No | Ag85A |
| B | 51790 | QPACRKAGCQTYKWE | Rv3804c | No | No | Ag85A |
| B | 70010 | VMPVGGQSSFYSDWY | Rv3804c | No | No | Ag85A |
| T | 1546 | AGGYKASDMWGPKEDPAWQR | Rv3804c | No | No | Ag85A |
| T | 3403 | ANRHVKPTGSAVVGLSMAAS | Rv3804c | No | No | Ag85A |
| T | 3422 | ANSPALYLLDGLRAQDDFSG | Rv3804c | No | No | Ag85A |
| T | 6901 | CQTYKWETF | Rv3804c | No | No | Ag85A |
| T | 8686 | DIKVQFQSGGANSPALYLLD | Rv3804c | No | No | Ag85A |
| T | 10838 | DWYQPACGKAGCQTYKWETF | Rv3804c | No | No | Ag85A |
| T | 18896 | GCQTYKWETFLTSELPGWLQ | Rv3804c | No | No | Ag85A |
| T | 19646 | GFVRTSNIKFQDAYNAGGGH | Rv3804c | No | No | Ag85A |
| T | 20979 | GLLDPSQAMGPTLIGLAMGD | Rv3804c | No | No | Ag85A |
| T | 21093 | GLRAQDDFSGWDINTPAFEW | Rv3804c | No | No | Ag85A |
| T | 21439 | GNGKPSDLGGNNLPAKFLEG | Rv3804c | No | No | Ag85A |
| T | 21482 | GNNLPAKFLEGFVRTSNIKF | Rv3804c | No | No | Ag85A |
| T | 21670 | GPKEDPAWQRNDPLLNVGKL | Rv3804c | No | No | Ag85A |
| T | 21960 | GQSSFYSDWY | Rv3804c | No | No | Ag85A |
| T | 25363 | IANNTRVWVYCGNGKPSDLG | Rv3804c | GCC-GGC | A-G | Ag85A |
| T | 31902 | KLIANNTRV | Rv3804c | GCC-GGC | A-G | Ag85A |
| T | 34823 | LAIYHPQQFVYAGAMSGLLD | Rv3804c | No | No | Ag85A |
| T | 40162 | LTSELPGWLQANRHVKPTGS | Rv3804c | No | No | Ag85A |
| T | 41872 | MKPDLQRALGATPNTGPAPQGA | Rv3804c | No | No | Ag85A |
| T | 43504 | NDPLLNVGKLIANNTRVWVY | Rv3804c | GCC-GGC | A-G | Ag85A |
| T | 44100 | NGVFDFPDSGTHSWEYWGAQ | Rv3804c | GCG-GGG | A-G | Ag85A |
| T | 49699 | PTLIGLAMGDAGGYKASDMW | Rv3804c | No | No | Ag85A |
| T | 49870 | PVGGQSSFYSDWYQPACGKA | Rv3804c | No | No | Ag85A |
| T | 50444 | QDAYNAGGGHNGVFDFPDSG | Rv3804c | No | No | Ag85A |
| T | 52030 | QQFVYAGAMSGLLDPSQAMG | Rv3804c | No | No | Ag85A |
| T | 52431 | QSSFYSDWY | Rv3804c | No | No | Ag85A |
| T | 56884 | SALTLAIYHPQQFVYAGAMS | Rv3804c | No | No | Ag85A |
| T | 64081 | THSWEYWGAQLNAMKPDLQR | Rv3804c | GCG-GGG | A-G | Ag85A |
| T | 72312 | WDINTPAFEWYDQSGLSVVM | Rv3804c | No | No | Ag85A |
| T | 73578 | YDQSGLSVVMPVGGQSSFYS | Rv3804c | No | No | Ag85A |
| T | 103416 | MQLVDRVRG | Rv3804c | GTT-GTC | No | Ag85A |
| T | 103423 | MSRRLVVGA | Rv3804c | No | No | Ag85A |
| T | 173920 | AMGPTLIGLAMGDAG | Rv3804c | No | No | Ag85A |
| T | 173921 | AMSGLLDPSQAMGPT | Rv3804c | No | No | Ag85A |
| T | 173924 | AYNAGGGHNGVFDFP | Rv3804c | No | No | Ag85A |
| T | 173925 | DSGTHSWEYWGAQLN | Rv3804c | GCG-GGG | A-G | Ag85A |
| T | 173926 | FEWYDQSGLSVVMPV | Rv3804c | No | No | Ag85A |
| T | 173927 | FLEGFVRTSNIKFQD | Rv3804c | No | No | Ag85A |
| T | 173932 | FSGWDINTPAFEWYD | Rv3804c | No | No | Ag85A |
| T | 173933 | FVYAGAMSGLLDPSQ | Rv3804c | No | No | Ag85A |
| T | 173934 | FYSDWYQPACGKAGC | Rv3804c | No | No | Ag85A |
| T | 173935 | GAQLNAMKPDLQRAL | Rv3804c | GCG-GGG | A-G | Ag85A |
| T | 173936 | GGQSSFYSDWYQPAC | Rv3804c | No | No | Ag85A |
| T | 173937 | GKAGCQTYKWETFLT | Rv3804c | No | No | Ag85A |
| T | 173938 | GKLIANNTRVWVYCG | Rv3804c | GCC-GGC | A-G | Ag85A |
| T | 173941 | KEDPAWQRNDPLLNV | Rv3804c | No | No | Ag85A |
| T | 173943 | KVQFQSGGANSPALY | Rv3804c | No | No | Ag85A |
| T | 173944 | LLDGLRAQDDFSGWD | Rv3804c | No | No | Ag85A |
| T | 173948 | MGRDIKVQFQSGGAN | Rv3804c | No | No | Ag85A |
| T | 173950 | NNTRVWVYCGNGKPS | Rv3804c | No | No | Ag85A |
| T | 173953 | PLLNVGKLIANNTRV | Rv3804c | GCC-GGC | A-G | Ag85A |
| T | 174003 | QTYKWETFLTSELPG | Rv3804c | No | No | Ag85A |
| T | 174004 | RAQDDFSGWDINTPA | Rv3804c | No | No | Ag85A |
| T | 174007 | SELPGWLQANRHVKP | Rv3804c | No | No | Ag85A |
| T | 174010 | SPALYLLDGLRAQDD | Rv3804c | No | No | Ag85A |
| T | 174014 | SWEYWGAQLNAMKPD | Rv3804c | GCG-GGG | A-G | Ag85A |
| T | 174016 | TGSAVVGLSMAASSA | Rv3804c | No | No | Ag85A |
| T | 174020 | VFDFPDSGTHSWEYW | Rv3804c | No | No | Ag85A |
| T | 174022 | WLQANRHVKPTGSAV | Rv3804c | No | No | Ag85A |
| T | 174023 | WQRNDPLLNVGKLIA | Rv3804c | GCC-GGC | A-G | Ag85A |
| T | 174024 | WVYCGNGKPSDLGGN | Rv3804c | No | No | Ag85A |
| T | 174025 | YHPQQFVYAGAMSGL | Rv3804c | No | No | Ag85A |
| T/B | 17838 | FSRPGLPVEYLQVPSPSMGR | Rv3804c | CCG-TCG CCG-CCC | P-S No | Ag85A |
| T/B | 39011 | LQVPSPSMGRDIKVQFQSGG | Rv3804c | No | No | Ag85A |
*The CDS of Ag85A, Ag85B and Ag85C of M. Tuberculosis H37Rv strain has been used as the reference sequence.
‡Bold and underlined AA indicates locations of amino acid changes.
Distribution of synonymous and nonsynonymous SNPs in gene sequence of Ag85A, Ag85B and Ag85C among 180 strains*
| Gene | Length (bp) | SNPs | dN | dS | dN/dS | |||
|---|---|---|---|---|---|---|---|---|
| Nonsyn | Syn | All | ||||||
| Ag85A | T cell epitope region | 939 | 3 | 2 | 5 | 0.000032 | 0.000190 | 0.17 |
| Non-T-cell-epitope region | 75 | 0 | 0 | 0 | 0 | 0 | 0 | |
| B cell epitope region | 315 | 1 | 1 | 2 | 0.000047 | 0.000146 | 0.32 | |
| Non-B-cell-epitope region | 699 | 2 | 1 | 3 | 0.000022 | 0.000183 | 0.12 | |
| All | 1014 | 3 | 2 | 5 | 0.000031 | 0.000134 | 0.23 | |
| Ag85B | T cell epitope region | 897 | 1 | 3 | 4 | 0.000034 | 0.002458 | 0.014 |
| Non-T-cell-epitope region | 78 | 1 | 0 | 1 | 0.000219 | 0 | NA | |
| B cell epitope region | 930 | 2 | 3 | 5 | 0.000049 | 0.002355 | 0.021 | |
| Non-B-cell-epitope region | 45 | 0 | 0 | 0 | 0 | 0 | NA | |
| All | 975 | 2 | 3 | 5 | 0.000047 | 0.002230 | 0.021 | |
| Ag85C | T cell epitope region | 96 | 0 | 0 | 0 | 0 | 0 | 0 |
| Non-T-cell-epitope region | 924 | 1 | 1 | 2 | 0.000122 | 0.000061 | 2 | |
| All | 1020 | 1 | 1 | 2 | 0.000108 | 0.000053 | 2.04 | |
* H37Rv was used as reference to base the change in allele for the SNPs
NA, not applicable
dN/dS values of proteins, epitope region and non-epitope region
Table 7 showed the distribution of synonymous and nonsynonymous SNPs in Ag85A, Ag85B and Ag85C among all 180 strains. The dN/dS value of Ag85A and Ag85B were 0.23 and 0.021, both much lower than 1, suggesting that these two proteins are likely appeared to be under purifying selection (Form of natural selection that acts to eliminate selectively deleterious mutations). For Ag85A, T/B cell epitope regions harbored higher dN/dS values than non-epitope regions, which mean the formers had accumulated significantly more amino acid changes than the latters. For Ag85B, non-T-cell-epitope regions had higher dN/dS value than T cell epitope regions, while B cell epitope regions owned higher dN/dS than non-B-cell-epitope regions. dN/dS of Ag85C was 2.04, higher than 1. All changes in Ag85C were from non epitope regions.
Changes in protein level
All of the M.bovis and BCG strains presented two unique mutations in A312G of Ag85A and F140L of Ag85B, which might represent special mutations in BCG strains. Six strains with mutation of G158S in Ag85C included four CAS strains (XZ06003, XJ06018, XJ06153 and XJ06188), one Beijing strain (FJ06159) and one new spoligotype strain (FJ05009). As the C714A in Ag85B showed high polymorphism, we counted the frequencies of the synonymous mutation (Table 7). Among the 92 isolates of Beijing genotype, 95 % (n = 87) of the isolates presented A; meanwhile, among the 88 non-Beijing isolates, only ten isolates were A in the position 714 of Ag85B.
Genetic diversity of antigens Ag85A, Ag85B and Ag85C among 180 strains. T/B cell epitope region are marked in the sequences.
DISCUSSION
In this study, we chose 180 clinical MTBC strains which were originated from a very large geographical area and have different spoligotyping patterns in China; hence the data provided by them could be representative of genetic diversity that might be present within China, at least to some extent.
Studies in human pathogenic viruses, bacteria and protozoa have revealed that genes encoding antigens tend to be highly variable as a consequence of diversifying selection to evade host immunity (19-22). Comas et al reported that human T cell epitopes of M. tuberculosis were evolutionarily hyperconserved and thus deduced that M. tuberculosis was lack of antigenic variation and immune evasion(3). However, in our previous studies, some proteins, such as MPT64, PstS1, Rv0309 and Rv2945c, harbored higher numbers of amino acid substitutions in their T cell epitopes, which suggesting their role in ongoing immune evasion (4,5,6). In this study, we found that Ag85A, Ag85B and Ag85C on the contrary showed lower substitution of amino acid in T/B cell epitopes. The dN/dS value of Ag85A and Ag85B were 0.23 and 0.021, both lower than 1, suggesting that these two proteins were likely appeared to be under purifying selection. dN/dS of Ag85C was 2.04, higher than 1. Yet all changes in Ag85C were from non epitope regions. Our data indicated that Ag85 were hyperconserved in T/B cell epitopes and the genes were more likely to be under purifying selection, which is in line with Comas' study.
Mycobacterium Ag85 complex consists of Ag85A, Ag85B, and Ag85C, which play an important role in cell wall biosynthesis by catalyzing the synthesis of the cord factor (trehalose 6,6′-dimycolate, TDM) by mycolyltransferase activity and serve as fibronectin-binding proteins that interact with host macrophage to trigger host immune response(23,24). M. tuberculosis secretes many proteins into the extracellular environment, which can be recognized by the host immune system and induce protective immunity and immune responses with diagnostic values. Our findings that Ag85 complex are highly conserved in T/B cell epitopes also indicate they are suitable for diagnose and vaccine for TB. Currently, there are several vaccine candidates undergoing clinical trials that represent vaccines with different immune profiles and modes of action. In a study, 2797 BCG-vaccinated infants were boosted with MVA expressing the MTB antigen 85A or a placebo control and thereafter followed for 3 years. However, the outcome of the trial was very disappointing with no detectable improvement of protection against TB (15). It was proposed that rather than boosting Th1 responses, we should focus on understanding protective immune responses that are lacking or insufficiently promoted by BCG that can intervene at critical stages of the TB life cycle (25).
Backus KM et al reported that the three Mycobacterium tuberculosis antigen 85 isoforms have unique substrates and activities determined by non-active site regions, which reflect the differences among the three antigens (26). Our study showed that each of these three antigens was conserved, suggesting similar activity and substrate selectivity among different strains.
Genes in M. tuberculosis was assigned functional categories according to Tuberculist (http://tuberculist.epfl.ch/). MPT64, PstS1, Rv2945c and Rv0309 all belong to virulence, detoxification, adaption proteins, while Ag85 are lipid metabolism proteins. Different functions between individual genes indicate they are under distinct selection pressures. The former category of proteins are the first proteins interact with host immune system after M. tuberculosis strains infected, which made them easier to be under host immune selection to induce immune evasion. However, Ag85 showed lower polymorphisms than the former proteins. The data showed that proteins Ag85 were under purifying selection. We assume that M. tuberculosis proteins in different categories are suffered divergence from host immune selection. More proteins should be included to clarify this point.
In position 312 of Ag85A, a nonpolar amino acid Ala was substituted by a polar one (Gly) in M.bovis and BCG strains, the hydrophilicity increased, which suggests that the protein is more likely to combine the membrane structures related to function. The mutation in F140L of Ag85B rarely affects antigen function since it was changed between two nonpolar amino acids, it could still be used as a good phylogenetic marker to differentiate M.bovis and BCG strains from M.tuberculosis strains. Six strains with mutation of G472A in Ag85C included four CAS strains (XZ06003, XJ06018, XJ06153 and XJ06188), one Beijing strain (FJ06159) and one new spoligotype strain (FJ05009). Therefore, the SNPs were acting as phylogenetic markers for the four CAS strains. For strains in different spoligotypes, i.e. non-closely related strains, we had one homoplastic SNP(convergent evolution), which usually is a strong indicator of selection. As the mutation were changed between two polar amino acids (G-A), it nearly did not affect the protein function. 95 % Beijing isolates presented A in position 714 of Ag85B, thus C714A in Ag85B seemed to be a valuable phylogenetic marker for Beijing strains.
In conclusion, proteins Ag85 are highly conserved in T/B cell epitopes and the genes are more likely to be under purifying selection. The divergence of host immune selection on different proteins may result from different function of the proteins. A312G of Ag85A and T418C of Ag85B may represent special mutations in BCG strains, which may be used to differentiate M.bovis and BCG strains from MTB strains.
Supplementary Material
Table S1.
Acknowledgements
We thank the staffs of the respective institutes in Beijing municipality, the 13 provinces and autonomous regions in China for their excellent contribution to this study, specially for the help of Lishui Zhang (Fujian), Yunhong Tan (Hunan), Xiujun Yang (Jilin), Chongxiang Tong (Gansu), Feiying Liu (Guangxi), Yingcheng Qi (Xinjiang), Qing Wang (Anhui), Xiaohui Cao and Ping Zhao (Beijing), Haitao Li (Henan), JunYang (Sichuan), Xuanmin Zhang (Shannxi), Li Shi (Xizang), Qing Wang (Anhui) and Xiaomeng Wang (Zhejiang).
Funding: This work was funded by the project 81401647 of Natural Science Foundation of China, 2013ZX10003006 and 2013ZX10003002-001of Chinese National Key Program of Mega Infectious Diseases.
Competing Interests
The authors have declared that no competing interest exists.
References
1. World Health Organization. Global tuberculosis report. 2012.
2. Young DB, Kaufmann SH, Hermans PW, Thole JE. Mycobacterial protein antigens: a compilation. Mol Microbiol. 1992Jan;6(2):133-145
3. Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010Jun;42(6):498-503
4. Jiang Y, Liu H, Wang H, Dou X, Zhao X, Bai Y, Wan L, Li G, Zhang W, Chen C, Wan K. Polymorphism of antigen MPT64 in Mycobacterium tuberculosis strains. J Clin Microbiol. 2013May;51(5):1558-62
5. Liu H, Jiang Y, Dou X, Wang H, Zhao X, Zhang W, Wan L, Zhang Z, Chen C, Wan L. pstS1 polymorphisms of Mycobacterium tuberculosis strains may reflect ongoing immune evasion. Tuberculosis (Edinb.). 2013Sep;93(5):475-81
6. Jiang Y, Dou X, Zhang W, Liu H, Zhao X, Wang H, Lian L, Yu Q, Zhang J, Li G, Chen C, Wan K. Genetic diversity of antigens Rv2945c and Rv0309 in Mycobacterium tuberculosis strains may reflect ongoing immune evasion. FEMS Microbiol Lett. 2013Oct;347(1):77-82
7. Wiker HG, Harboe M. The antigen 85 complex: a major secretion product of Mycobacterium tuberculosis. Microbiol Rev. 1992Dec;56(4):648-61
8. Armitige LY, Jagannath C, Wanger AR, Norris SJ. Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv: effect on growth in culture and in macrophages. Infect Immun. 2000Feb;68(2):767-78
9. Elamin AA, Stehr M, Oehlmann W, Singh M. The mycolyltransferase 85A, a putative drug target of Mycobacterium tuberculosis: development of a novel assay and quantification of glycolipid-status of the mycobacterial cell wall. J Microbiol Methods. 2009Dec;79(3):358-63
10. Kamath AT, Rochat AF, Valenti MP, Agger EM, Lingnau K, Andersen P, Lambert P, Siegrist CA. Adult-like antimycobacterial T cell and in vivo dendritic cell responses following neonatal immunization with Ag85B-ESAT-6 in the IC31 adjuvant. PLoS One. 2008;3(11):e3683
11. Andersen P, Doherty TM. The success and failure of BCG—implications for a novel tuberculosis vaccine. Nat Rev Microbiol. 2005Aug;3(8):656-62
12. Doherty TM, Olsen AW, Weischenfeldt J, Huygen K, D'Souza S, Kondratieva TK, Yeremeev VV, Apt A, Raupach B, Grode Leander, Kaufmann S, Andersen P. Comparative analysis of different vaccine constructs expressing defined antigens from Mycobacterium tuberculosis. J Infect Dis. 2004Dec;190(12):2146-53
13. Weinrich Olsen A, van Pinxteren LA, Meng Okkels L, Birk Rasmussen P, Andersen P. Protection of mice with a tuberculosis subunit vaccine based on a fusion protein of antigen 85b and esat-6. Infect Immun. 2001May;69(5):2773-8
14. Langermans JA, Doherty TM, Vervenne RA, van der Laan T, Lyashchenko K, Greenwald R, Aagaard C, Weiler Horst, Soolingen DV, Dalemans W. Protection of macaques against Mycobacterium tuberculosis infection by a subunit vaccine based on a fusion protein of antigen 85B and ESAT-6. Vaccine. 2005Apr15;23(21):2740-50
15. Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, Shea JE, McClain JB, Hussey GD, Hanekom WA, Mahomed H, McShane H, MVA85A 020 Trial Study Team. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet. 2013Mar23;381(9871):1021-8
16. Dong H, Liu Z, Lv B, Zhang Y, Liu J, Zhao X, Wan K. Spoligotypes of Mycobacterium tuberculosis from different Provinces of China. J Clin Microbiol. 2010Nov;48(11):4102-4106
17. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007Nov1;23(21):2947-8
18. Ernst JD, Lewinsohn DM, Behar S, Blythe M, Schlesinger LS, Kornfeld H, Sette A. Meeting Report: NIH Workshop on the Tuberculosis Immune Epitope Database. Tuberculosis (Edinb.). 2008Jul;88(4):366-370
19. Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung'u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009Apr2;458(7238):641-645
20. Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000Apr14;288(5464):339-344
21. Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, Kyes S, Krishna S, Newbold C, Dermitzakis ET, Berriman M. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007Jan;39(1):120-5
22. Urwin R, Russell JE, Thompson EA, Holmes EC, Feavers IM, Maiden MC. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect Immun. 2004Oct;72(10):5955-62
23. Ohara N, Nishiyama T, Ohara-Wada N, Matsumoto S, Matsuo T, Yamada T. Characterization of the transcriptional initiation regions of genes for the major secreted protein antigens 85C and MPB51 of Mycobacterium bovis BCG. Microb Pathog. 1997Nov;23(5):303-10
24. Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997May30;276(5317):1420-2
25. Andersen P, Woodworth JS. Tuberculosis vaccines--rethinking the current paradigm. Trends Immunol. 2014Aug;35(8):387-95
26. Backus KM, Dolan MA, Barry CS, Joe M, McPhie P, Boshoff HI, Lowary TL, Davis BG, Barry CE 3rd. The Three Mycobacterium tuberculosis antigen 85 isoforms have unique substrates and activities determined by non-active site regions. J Biol Chem. 2014Sep5;289(36):25041-53
Author contact
![]() Corresponding author: National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, P. O. Box 5, Changping, Beijing 102206, People's Republic of China. Phone and fax: 0086 10 58900779. E-mail: wankanglincn
Corresponding author: National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, P. O. Box 5, Changping, Beijing 102206, People's Republic of China. Phone and fax: 0086 10 58900779. E-mail: wankanglincn

 Global reach, higher impact
Global reach, higher impact