3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(1):32-41. doi:10.7150/ijms.9840 This issue Cite
Research Paper
A Microsatellite Polymorphism in IGF1 Gene Promoter and Timing of Natural Menopause in Caucasian Women
1. Department of Human Biological Development, Institute of Anthropology, Faculty of Biology, Adam Mickiewicz University in Poznań, Poland
2. Department of Animal Physiology, Institute of Experimental Biology, Faculty of Biology, Adam Mickiewicz University in Poznań, Poland
3. Department of Gynaecological Oncology and Gynaecology, Medical University, Lublin, Poland
4. Department of Obstetrics, Endocrinology and Gynaecology, Poznań University of Medical Sciences, Poland
5. Department of Molecular Virology, Institute of Experimental Biology, Faculty of Biology, Adam Mickiewicz University in Poznań, Poland
Received 2014-6-7; Accepted 2014-8-21; Published 2015-1-1
Abstract
Background: Genes involved in the IGF-1 aging pathways in the human ovary can be considered strong candidates for predictors of the natural menopause timing. This study evaluates the association between a cytosine-adenine (CA) microsatellite polymorphism in the IGF1 gene promoter P1 and age at natural menopause.
Methods: Genomic DNA was extracted from the peripheral blood, PCR was performed using primers designed to amplify the polymorphic (CA)n repeat of the human IGF1 gene, an allele dose effect for the most common (CA)19 repeats allele, Cox proportional hazard regression models and the Kaplan-Meier cumulative survivorship method with the log-rank test were used to determine statistical significance of studied associations in a sample of 257 Polish women aged 40-58 years.
Results: Crude Cox proportional hazard regression analysis confirmed the association between the IGF1 gene polymorphism and the menopause timing (p=0.038). This relationship remained statistically significant after controlling for other menopause confounders in multivariate modelling. Out of the input variables, the (CA)n polymorphism in the IGF1 gene promoter, age at menarche and smoking status were independent covariates of the natural menopause timing (χ2 =12.845; df=3; p=0.034). The onset of menopause at a younger age was likely associated with the IGF1 genotype variant not carrying the (CA)19 repeats allele, menarche before the age of 12 and a current cigarette smoker status (HR=1.6).
Conclusion: This study provides evidence that a common cytosine-adenine (CA) microsatellite repeat polymorphism in the P1 promoter region of the IGF1 gene is an independent predictive factor for age at natural menopause in Caucasian women also after adjusting for other menopause covariates.
Keywords: candidate gene approach, cytosine-adenine (CA) microsatellite polymorphism, IGF1 gene, age at natural menopause, age at menarche, smoking status
Introduction
Menopause is a life-history trait that marks the end of a woman's reproductive life and uniquely features her life cycle typically occurring well before senescence of other physiological functions and long before reaching maximum life expectancy.1 Physiologically, menopause is characterized by a permanent cessation of menstruation resulting from continuous depletion of a fixed number of primordial follicles, the great majority of which are lost due to atresia, leading to almost total depletion and loss of ovarian follicular activity at the female mid-life. Depletion in follicle supply is accompanied by a decline of circulating estrogen and its metabolites and a relative increase of circulating androgens causing typical vasomotor, somatic, urogenital and psychological alterations.2
This curtailing of female reproduction commonly occurs between 45 and 55 years of age with worldwide estimates of average age at natural menopause varying from 44.6 years in Indian women from Punjab, 51.2 years in Polish and 52.0 years in French women.3,4 There has been evidence of shifting the natural menopause timing across Europe towards older ages, thereby confirming suggestions for secular trends.5
In recent years, the timing of menopause has gained attention for several reasons. Firstly, in most Western countries, a trend towards delayed maternal first birth has been recognized. This new trend is generally associated with a reduction in female total fertility. In this context, prediction of menopause timing, i.e. the onset of subfertility and/or infertility, is of clinical significance.6 Secondly, the menopause timing is likely to be associated with common diseases. There is epidemiologic evidence that an early onset of menopause is associated with an increased risk of cardiovascular diseases, osteoporosis and overall mortality, whereas a later age at menopause is associated with an elevated risk of breast, ovarian, and endometrial cancers.7-9
Age at menopause may be considered a complex quantitative trait that is determined by multiple factors: genetic, vascular and environmental. Population, twin and family-based studies suggest that the wide age range for natural menopause is most likely due to gene-environment interactions of many genes with widespread environmental exposures over the female lifespan. The heritability estimates range from 31 to 78%.10-12 Environmental factors affecting the onset of menopause are commonly attributed to life-history traits; specifically to characteristics of the individual's reproductive period, socio-economic status and lifestyle behaviour, and they are likely to account only for a small part (ca 4%) of the total menopause age variation.4,13,14
Recent genome-wide linkage (GWL) analyses using microsatellite markers have identified chromosomal regions likely to store genes for the onset of natural menopause.10,11,15
Some novel genetic loci associated with menopausal age have been identified within genome wide association (GWAS) studies. The recent discovery of thirteen new regions of the genome associated with menopause timing has shed new light on the biological pathways involved in the processes of reproductive ageing and age-related diseases.16 To date, more than 17 novel genetic loci have been identified in GWAS for age at natural menopause.17
In a 2010 large-scale candidate-gene association study of age at menarche and age at natural menopause, He and her colleagues proposed eight groups of candidate genes that may play a role in natural menopause age; including genes involved in biologically plausible pathways and those for related phenotypes.18 They also revealed a statistically significant association at the gene level between the IGF1 gene and natural menopause age.
The IGF1 gene has a single copy in the human genome located on chromosome 12 (12q22.1-q24.1).19 The genomic sequence is approximately 85,000 bases long and has 6 exons.20 The IGF1 gene transcription is controlled by two promoters referred to as P1 and P2 and situated before exons 1 and 2, respectively.21 The main promoter region P1 consists of the DNA fragment situated upstream from the transcription start site, 1,630 base pairs long and a fragment of exon 1, the so-called 5'UTR, 322 base pairs long. The exon 1 5'UTR fragment is indispensable for regular and efficient transcription of this gene and remains its most conserved part.22 The most common analyzed polymorphism in IGF1, located in the 5' regulatory region of the gene, is a cytosine-adenine (CA) microsatellite polymorphism, 969 kbp upstream from the IGF1 gene promoter.23 It comprises a variable length of CA repeats ranging from a minimum of 10 to a maximum of 23 repeats; the most common allele has 19 repeats (CA19).24,25 This polymorphism is thought to alter promoter activity and thus influence the transcription rate of IGF1 but conclusive proof of this association has not yet been established.26,27
The IGF-1 signaling pathway has characteristics of both a circulating hormone released from the liver in response to stimulation by GH and a tissue growth factor and can therefore expand its effects through autocrine or paracrine mechanisms. The mitogenic and antiapoptotic actions of IGF-1 are essential for normal growth throughout foetal and childhood development. In adult life, however, aberrant stimulation can lead to excessive proliferation and survival signals and the development of different epithelial tumor types.28
Gene knockout experiments in mice have demonstrated that the IGF-1 axis is required for a normal rate of sexual development and maturation, and ovarian function; the IGF-1 knockout mice have been reported to be sterile.29 There is evidence that IGF-1 has a stimulatory effect on ovarian function including (i) follicular cell replication by activating growth and differentiation of the ovarian follicles, development of preantral follicles, maintaining the larger pool of small antral follicles, stimulating the development of follicles, and selecting dominant follicle; and (ii) steroidogenesis in theca cells and secretion of progesterone by large antral follicles and gonadotropin action.30
Genes involved in the IGF-1 biological pathway, particularly in ovarian ageing, can be viewed as candidate genes for the timing of menopause. On the basis of this presumption, the study aims to test whether a cytosine-adenine (CA) microsatellite polymorphism in the IGF1 gene promoter is associated with the timing of natural menopause in Caucasian women either as a single predictor or after adjusting for other possible menopause risk factors.
Materials and Methods
Study design and participants
This study was a cross-sectional survey and was conducted between January and April 2011. Participants were 257 women between the ages of 40 and 58 years. They were recruited from the Women's Reproductive Health Programme implemented at the Adam Mickiewicz University in Poznań in cooperation with Poznań University of Medical Sciences and Medical University of Lublin. The eligibility criteria for this study were restricted to a cohort of women aged between 40 and 60 years, without a history of cancer, surgical menopause or endocrine system diseases, not pregnant, not on hormonal replacement therapy (HRT) and not using insulin. This research project was reviewed and approved by the Bioethics Committee in Poznań University of Medical Sciences and was carried out in accordance with the Declaration of Helsinki and subsequent amendments. All participants indicated informed consent by signing a form after they had been given a clear explanation of the study objectives and potential risks of the study.
The study protocol included a medical examination, collection of blood samples and completion of an MSQ-questionnaire.
Interest focused on the assessment of the IGF1 gene promoter polymorphism length, past reproductive events and the lifestyle behaviours hypothesized as covariates of age at natural menopause. The Polish version of menopause-specific questionnaire (MSQ) was administered to study participants and self-reported socio-demographic, lifestyle behaviour and reproductive history data were collected.31 The natural menopause was referred to the last menstrual period (LMP) in a woman's life resulting from cessation of ovarian follicular activity. The questions regarding menopausal status used both status-quo and retrospective techniques. The first question asked “Have your menstrual periods ceased permanently?” If yes, “How much time has passed since the last menstrual period? “ Response categories were: “3 months, 6 months, 12 months, more than 12 months”. The next question was “At what age did your natural periods cease?” Women recalled their age at the last menstrual period (LMP) and the elapsed time. This information was taken to check the reliability of women's responses because the status-quo approach was used in all statistical analyses considering menopausal age. On the basis of the responses, the study participants were then stratified by menstrual status using WHO recommendations.32 The premenopausal status considered ongoing and regularly menstruating women. The postmenopausal status was defined as amenorrhea for 12 or more months dating from the last menstrual period, for which there were no other obvious pathological or physiological causes for menses cessation.
Genotype assays and genotyping
The participants donated blood samples at the time of medical examination. Nurses drew fifteen millimeters of blood. Fasting blood samples were obtained during morning hours by venipuncture and stored in EDTA-3K vacuum tubes to prevent coagulation. Serum samples were separated according to a standardized protocol by low speed centrifugation for 10 min at 3000 rpm at room temperature. Serum, buffy coat, and red cells were then aliquoted into cryovials and stored deep-frozen at -80oC until use for genetic analyses. Genomic DNA was isolated from blood using the Blood Mini Kit (A&A Biotechnology, Poland) according to the manufacturer's protocol, and data was thus obtained from 257 individuals.
PCR was performed using primers designed to amplify the polymorphic (CA)n repeat of the human IGFI gene. The reaction was carried out in a final volume of 15 ml, containing 100 ng genomic DNA, 3.75 pmol forward primer (5'-GAAAACACACTCTGGCAC-3') labelled with FAM, 3.75 pmol reverse primer (5'-ACCACTCTGGGAGAAGGGTA-3'), 0.1 mM deoxy-NTP, 1.5 mM MgCl2, 1X PCR buffer and 0.6 U HiFi DNA Polymerase (Novazym, Poland, Cat. no. N1003-05). PCR was performed using a thermal cycler (Tgradient Thermocycler, Biometra, Germany) with the parameters: 94o C for 4 min; 28 PCR cycles of 5 sec. at 94o C, 30 sec. at 60o C and completed with a final extension for 30 min. at 65o C. Analysis of PCR product sizes was performed on an automated sequencing apparatus (ABI 3130xl) and determined in comparison with the internal GS600LIZ size standard (Applied Biosystems). The estimation of CA repeat numbers in each analyzed specimen was based on extrapolation to a previously developed specific allelic ladder. The ladder marker consisted of 14 sequenced amplicons representing alleles with 7, 9, 11, 13 and 23 CA repeats.
Variables and statistical analyses
The dependent variable was age at natural menopause and the independent variables were its hypothesized genetic and non-genetic covariates. Subjects were first categorized in 17 genotypes based on 7 different alleles that were present in the sample and then allocated to genotype groups depending on the presence of the most frequent (CA)19 repeat allele: homozygotes with the 19CA repeats (CA)19/(CA)19 as the reference category, heterozygotes carrying one copy of the (CA)19 repeats (CA)19/(CA)non-19 and as the “others” category for those not carrying any copies of the (CA)19 repeats (CA)non-19/(CA)non-19. Chi-square tests assessed whether IGF1 genotype frequencies fell within the Hardy-Weinberg equilibrium. An allele dose effect was used for evaluating the hypothesized association of IGF1 gene promoter polymorphism and age at natural menopause: (1) homozygous (CA)19/(CA)19 versus (2) heterozygous (CA)19/(CA)non-19 and versus (3) non-carriers of 19 CA repeats.
Selected biological (life-history traits), socio-demographic and lifestyle variables hypothesized to be associated with menopause age included: 1) age at menarche, reported as age at first menstrual period, ranging from 11 (referent) to 17 years; 2) age at first childbirth among parous women with at least one live birth, ranging from 16 (referent) to 33 years; 3) parity, defined as the number of live births ever given by a woman was analyzed as dichotomous variable (nulliparous versus parous) and as number of live births (none - referent, 1, 2, 3 and more); 4) use of hormonal contraceptives (OCU) (never used - referent, ever used, for at least one year) was analyzed as dichotomous variable; 5) weight status on the basis of BMI, calculated as the weight in kilograms divided by the square of the height in meters (kg/m2), ranging from 19.6 kg/m2 (reference) to 41.2 kg/m2; 6) marital status with married/cohabitating as referent, formerly married and never married; 7) educational attainment: primary/vocational<12 years - referent, secondary =12 years, and tertiary>12 years; 8) smoking status at blood donation: never smoked - referent, past smoker, smoking at baseline; 9) general health status at baseline was self-rated on a five-point Likert-type scale ranging from “poor“ (referent) to “good, excellent, could not be better” and was analyzed as a dichotomous variable less than good - referent versus good or very good.
Data were analysed using the discrete-time method for analysis of events. Univariate Cox proportional hazards regression analysis was used to evaluate the crude effect of the IGF1 gene promoter polymorphism and other potential non-genetic covariates on the timing of menopause. The proportionality assumption was confirmed by visual inspection, plotting the Kaplan-Meier survival curves. The Wald test was used to determine statistical significance of explanatory variables and was defined by a p-value of less than 0.05.
Multivariate Cox proportional hazard regression analysis was used to estimate the independent effects of the polymorphisms after controlling for non-genetic exposure variables. Only those variables that had been significantly associated with age at menopause in bivariate relationships were included in the initial multivariate model. Using a backwards elimination approach, the final model was obtained after dropping in a step-wise fashion variables that had not significantly influenced the model. The probability threshold for removal was set at 0.05.
Results of the proportional hazard regression models were expressed as hazard ratios (HR) and 95% confidence intervals (CI). For ease of interpretation, the HRs of continuous variables were presented by their categories. The risk of menopause in each category was compared to the referent category. Relevant relative risk for earlier age at menopause was assessed and a p value for linear trend across categories was calculated using the likelihood ratio Chi-square test.
Median age at natural menopause was estimated using the Kaplan-Meier cumulative survivorship method with the log-rank test used to determine statistical significance. For survival analysis, the endpoint age was defined as one of the following 1) age at natural menopause, i.e. age at LMP followed by 12 months of amenorrhea, 2) age when bleeding had stopped for between 3 and 12 months, 3) current age if a woman was still menstruating. All but age at natural menopause were considered censored observations.
Statistical analyses were performed by the STATISTICA 10.0 data analysis software system (Stat Soft, Inc. Tulsa, OK USA). All significance tests comprised two-way determinations. A value of p<0.05 was considered statistically significant.
Results
The background characteristics are shown in Table 1.
Mean age of women enrolled in the study was 53.8 years, SD=4.2 years. Forty-seven percent of them were still menstruating (mean age 40.8 years; SD=4.2 years) and 52.5% were postmenopausal (mean age 55.1 years; SD=4.1 years). The Kaplan-Meier estimate of overall median age at menopause was 51.6 years (Q1=52 and Q3=54 years). Mean age at menarche was 13.0 years; SD=1.3 years. Mean age at first live birth given by a woman was 25.2 years; SD=4.3 years. Married/cohabitating women with tertiary level of education and two children were accounted for majority of the sample (83.1%, 63.8% and 40.1%). Every third woman reported the use of hormonal contraceptive pills (36.3%) and every fourth reported being a current smoker (25.3%). The proportion of those who rated their health as lower than good was 63.1% of the sample.
Background characteristics of women enrolled in the study.
| Characteristic | Value (n=257) | |
|---|---|---|
| Age (years) | 53.8±4.2a | |
| Age at menarche (years) | 13.0±1.3 | |
| Age at first live birth, parous only (years) | 25.2±4.3 | |
| BMI (kg/m2) | 26.4±3.7 | |
| Menstrual status | ||
| Premenopausal | 122 | (47.5)b |
| Postmenopausal | 135 | (52.5) |
| Marital status | ||
| Married/Cohabitating | 214 | (83.1) |
| Formerly Married/Cohabitating | 32 | (12.3) |
| Never Married/Cohabitating | 11 | (4.6) |
| Educational attainment | ||
| Primary/vocational <12 years | 38 | (14.8) |
| Secondary 12 years | 55 | (21.4) |
| Tertiary >12 years | 164 | (63.8) |
| Past OCUc | 95 | (36.9) |
| Parity, number of live births | ||
| Nulliparous | 32 | (12.4) |
| Parous 1 | 85 | (33.1) |
| 2 | 103 | (40.1) |
| ≥3 | 37 | (14.4) |
| Smoking status | ||
| Never smoked | 126 | (49.0) |
| Past smoker | 66 | (25.7) |
| Current smoker | 65 | (25.3) |
| Self-Rated Health | ||
| Less than good | 162 | (63.1) |
| Good or very good | 95 | (36.9) |
a Values are given as mean±standard variation.
bValues are given as number (percentage).
cOCU - Use of oral contraceptives for at least one year in the past
The 5'-end of the IGF-1 gene was flanked by a microsatellite marker of cytosine adenine (CA)n repeats. A total of seven alleles containing 11 to 22 CA repeats were identified in the sample (Table 2). Allele frequencies stratified by menopausal status were similar in order and magnitude to combined frequencies (p=0.54), with the (CA)19 repeat allele being the most frequent (63.5%). Two other variant types of alleles, the (CA)20 repeats (15.1%) and the (CA)21 repeats (10.4%) were next in order of frequency. The (CA)18 repeat allele was found at 8.5%, and the frequency of others, including alleles carrying 11, 17 and 22 CA repeats totaled less than 1%.
The most common genotype for the IGF1 polymorphism was homozygous for the (CA)19 repeats (19/19) and this accounted for 38.5% of the sample (Table 2). Other subjects were heterozygous. Frequency of heterozygotes carrying the (CA)19 repeat was 16.2% of the 19/20, followed by the 19/21 (16.6%), 10.5% of the 18/19 and 1.6% of all other 19/non-19 CA repeats. Other genotypes were identified in 13 variant types not carrying the 19 CA repeat allele. Each of their frequencies was less than 1%, and these were combined in one group labelled “others”; with a total frequency of 10.1%. The genotype distribution fell within the Hardy-Weinberg equilibrium (p=0.06).
Bivariate Cox proportional hazards regression models revealed the association of age at natural menopause with IGF1 (CA)n genotype variants (p=0.038), age at menarche (p=0.041), age at first live birth delivered by a woman (p=0.046), parity (p=0.050), and smoking status (p=0.013). The following variables: marital status, educational attainment, prior use of oral contraceptives, weight status (BMI), and perceived health status were not associated with menopause timing (Table 3).
All variables associated with menopausal age in the univariate analyses at p<0.5 significance were eligible for inclusion in the multivariate Cox proportional hazard regression model (Table 4).
Percentage distribution of the IGF1 cytosine-adenine dinucleotide repeats in the study population.
| Sample | IGF-1 cytosine-adenine repeats (CA)na (%) | IGF1 (CA)n genotype variants (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 17 | 18 | 19 | 20 | 21 | 22 | 19/19 | 18/19 | 19/20 | 19/21 | 20/20 | Otherb | |
| Premenop n=122 | 0.62 | 0.62 | 9.62 | 60.49 | 17.90 | 10.49 | 0.62 | 36.80 | 10.31 | 17.81 | 14.90 | 10.21 | 9.97 |
| Postmenop n=135 | 0.88 | 0.88 | 7.89 | 65.79 | 11.40 | 11.40 | 0 | 38.21 | 10.71 | 16.11 | 16.22 | 7.76 | 10.99 |
| p-value=0.54c | p-value=0.63 | ||||||||||||
| Total n=257 | 0.64 | 1.31 | 8.55 | 63.49 | 15.15 | 10.53 | 0.33 | 38.46 | 10.53 | 16.19 | 16.59 | 8.09 | 10.12 |
aNumber of alleles per person.
b All other non-carrier (CA)19
cχ2 Pearson's p-value
Univariate Cox proportional hazard regression of age at natural menopause on IGF1 (CA)n genotypes and selected biological, socio-demographic and lifestyle factors.
| N=257 Variable | ß | SE | Wald χ2 | HR (95%CI) | p |
|---|---|---|---|---|---|
| IGF1 (CA)n genotypes | 0.390 | 0.127 | 4.005 | 1.331 (0.152;1.891) | 0.038 |
| Age at menarche (years) | -0.138 | 0.078 | 3.698 | 0.870 (0.568;1.334) | 0.041 |
| Age at first live birtha (years) | -0.072 | 0.036 | 3.987 | 0.929 (0.865;0.998) | 0.046 |
| Parity, number of live births | -0.267 | 0.141 | 3.577 | 1.306 (0.990;1.723) | 0.050 |
| Marital status | 0.134 | 0.249 | 0.287 | 1.143 (0.700;1.866) | 0.592 |
| Educational attainment (years) | -0.118 | 0.183 | 0.418 | 0.888 (0.621;1.271) | 0.518 |
| Oral Contraceptives Use | -0.263 | 0.184 | 1.085 | 0.769 (0.623;1.005) | 0.089 |
| BMI (kg/m2) | 0.022 | 0.032 | 0.530 | 1.023 (0.962;1.087) | 0.466 |
| Smoking status | 0.657 | 0.267 | 6.049 | 1.929 (1.142;3.259) | 0.013 |
| Self-rated health | 0.130 | 0.190 | 0.470 | 1.139 (0.785;1.654) | 0.493 |
a Only parous women
Multivariate Cox proportional hazard regression models of age at natural menopause and the IGF1 (CA)n genotype variants and other potential covariates.
| Variable | n | Initial | Finala | Median age at menopauseb (Q1; Q3) |
|---|---|---|---|---|
| model HR (95%CI)c | model HR (95%CI)d | |||
| IGF1 (CA)n genotypes | ||||
| Homozygous (CA)19 (ref.) | 99 | 1 | 1 | 52.5 (50.5; 54.0) |
| Heterozygous (CA)19 | 123 | 1.29 (1.06;1.58) | 1.32 (1.09;1.61) | 51.7 (50.0; 53.0) |
| Non-carrier (CA)19 | 35 | 1.68 (1.38;2.05) | 1.75 (1.44;2.12) | 50.0 (47.1; 52.0) |
| p-value for trend | 0.042 | 0.040 | 19/19 vs non-19 p=0.027 | |
| 19/other vs non-19 p=0.038 | ||||
| Age at menarche (years) | ||||
| <12 (ref.) | 69 | 1 | 1 | 50.0 (47.0; 52.5) |
| 12 - 14 | 124 | 0.87 (0.75;1.01) | 0.80 (0.69;0.97) | 51.5 (50.0; 54.0) |
| >14 | 64 | 0.76 (0.65;0.89) | 0.64 (0.49;0.84) | 52.8 (52.0; 55.0) |
| p-value for trend | 0.038 | 0.030 | <12 vs >14 p=0.021 | |
| Age at first live birth (years) | ||||
| <20 (ref.) | 20 | 1 | ||
| 20-24 | 85 | 0.89 (0.75;1.07) | ||
| 25-29 | 60 | 0.80 (0.67;0.96) | ||
| ≥30 | 60 | 0.72 (0.60;0.86) | ||
| p-value for trend | 0.238 | |||
| Number of live births | ||||
| Nulliparous (ref.) | 32 | 1 | ||
| 1 | 84 | 0.72 (0.58;1.00) | ||
| 2 | 103 | 0.68 (0.54;0.97) | ||
| ≥3 | 38 | 0.59 (0.44;0.79) | ||
| p-value for trend | 0.219 | |||
| Smoking status | ||||
| Never (ref.) | 127 | 1 | 1 | 52.8 (51.0; 55.0) |
| Past | 65 | 1.74 (0.90;3.37) | 1.80 (0.93;3.48) | 52.0 (50.0; 53.0) |
| Current | 65 | 3.00 (1.55;5.81) | 3.25 (1.68;6,26) | 50.0 (49.0; 52.0) |
| p-value for trend | 0.024 | 0.018 | Never vs Current p=0.028 |
a The final model was obtained using stepwise backward procedure. Multivariate Cox proportional hazards regression model n=257; χ2 =12,845; df=3; p=0.034 adjusted for IGF1 (CA)n genotypes, age at menarche, and smoking status.
bMedian age calculated using Kaplan-Meier estimates; Q1 and Q3.
cHR(95%CI) - Crude hazard ratio and 95% confidence interval.
dHR(95%CI) - Adjusted hazard ratio and 95% confidence interval for IGF-1 (CA)n genotypes, age at menarche, and smoking status.
The initial multivariate Cox proportional hazards regression model involved the following covariates of menopause: the IGF1 genotype variants, age at menarche, age at first live birth, parity and smoking status. In the stepwise backward procedure, age at first live birth and parity lost their predictive value for age at menopause and the final model consisted of the IGF1 (CA)n genotype variants, age at menarche and smoking status (in the sample of 257 women, χ2 =12.845; df=3; p=0.034; a p-value for trend was 0.04, 0.03 and 0.018 for each variable, respectively).
The Kaplan-Meier survival curves for age at menopause stratified by the three IGF1 (CA)n genotype variants are shown in Figure 1.
The median menopause age of subjects homozygous for the (CA)19 repeats was 52.5 years (Q1=50.5; Q3=54 years), while of those heterozygous for the (CA)19 repeats (19/non-19 CA repeats) was 51.7 years (Q1=50; Q3 =53 years) and those not carrying (CA)19 repeats it was 50 years (Q1=47.1; Q3=52 years). The log-rank test revealed a trend in menopausal age across three IGF1 genotype variant types (p=0.04). In pairwise comparisons, subjects carrying the (CA)19 repeats as homozygous or heterozygous genotypes revealed a statistically significant difference to those not carrying the (CA)19 repeats (19/19 versus non-19/non-19; p=0.027) and 19/other versus non-19/non-19 (p=0.038). Homozygous carriers of the (CA)19 repeat allele were likely to experience menopause 2.5 years later than their counterparts not carrying this allele, while heterozygous (CA)19 carriers commenced as many as 1.7 years later than those without the (CA)19 repeat allele (Table 4).
Adjusted survival curves for age at natural menopause according to IGF1 genotype variants: 1. Homozygotes (CA)19; 2. Heterozygotes 19/non-19; 3. Non-carriers of (CA)19, adjusted for age at menarche and smoking status.
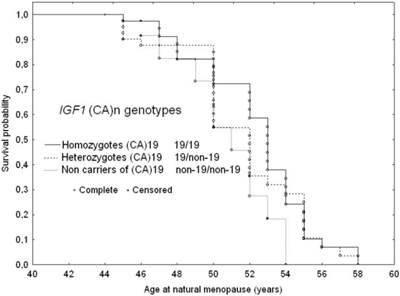
There was a trend of increasing hazard for earlier menopause onset with the absence of the 19 CA repeats. In subjects not carrying the (CA)19 repeat allele the risk of beginning menopause at a younger age was almost twice as high as in those homozygous for 19 CA repeats. Heterozygous carriers of (CA)19 repeat allele were likely to be at a 1.3 fold higher risks for earlier menopause than their homozygous for 19 CA repeats counterparts (Table 4 and Figure 1).
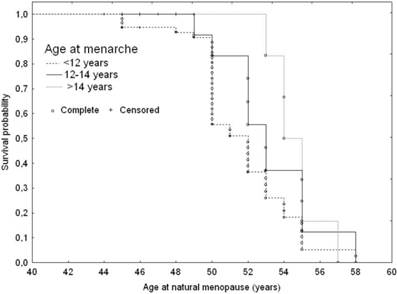
Age at menarche was directly associated with age at menopause (Table 4 and Figure 2). Late maturing subjects were likely to be at a 1.6 fold lower risk for earlier menopause onset than those maturing early. The median menopause age of subjects with menarche prior to 12 years of age was 50 years (Q1=47; Q3=52.5 years), while the late maturing women were likely to experience menopausal transition 2.8 years later than the early maturers (52.8; Q1=50; Q3=55 years; p=0.028).
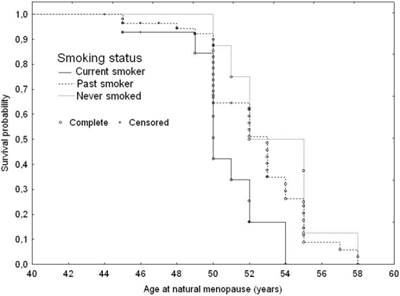
Currently smoking subjects were likely to be at a three-fold higher early menopausal risk (Table 4 and Figure 3). They were likely to experience menopause 2.8 years earlier than those who never smoked (50; Q1=49; Q3=52 years versus 52.8; Q1=51; Q3=55 years p=0.028).
Adjusted ssurvival curves for age at natural menopause according to age at menarche: 1. Younger than 12 years; 2. 12-14 years; 3. Older than 14 years, adjusted for IGF1 genotype variants and smoking status.
Adjusted survival curves for age at natural menopause according to smoking status: 1. Current smoker; 2. Past smoker; 3. Never smoked, adjusted for IGF1 genotype variants and age at menarche.
Discussion
This explorative study estimates the association between a common cytosine-adenine (CA) microsatellite polymorphism in the IGF1 gene and the timing of natural menopause in Caucasian women. In our study sample, seven alleles ranging in size from 11 to 22 CA repeats were identified in the promoter region of the IGF1 gene. Theoretically, it is possible to find 14 different alleles as the repeat lengths vary from a minimum of 10 repeats to a maximum of 23 repeats but 7 different alleles have also been found in another sample of Caucasian women.33 Mainly the (CA)19 and (CA)20 repeat alleles were present in the sample. The allele frequency of the (CA)19 repeats was 63.5% (60.5% in pre- and 65.8% in postmenopausal women), which corresponds to the allele frequency reported in other Caucasian populations, estimated to be between 59% and 70%.34 The allele frequency we found for the (CA)20 repeats was 15.1% (17.95% in pre- and 11,4% in postmenopausal women) and was consistent with other studies.26,34,35 The most common genotype variant was homozygous carrier of the (CA)19 repeats (38.1%) followed by heterozygous (CA)19/(CA)20 found in 20.9% subjects in our sample. Both data are in line with former reports of other Caucasian populations estimated to be between 37.3% and 46.8%.26,33-36
With respect to menopause covariates, a crude Cox proportional hazard regression analysis confirmed the association between the IGF1 gene cytosine-adenine (CA) microsatellite polymorphism and the menopause timing. Furthermore, we found that this relationship remained statistically significant after adjusting for other exposure variables in multivariate modelling. Out of the input variables, the (CA)n polymorphism in the IGF1 gene promoter, age at menarche and smoking status were independent covariates of the natural menopause timing. The onset of menopause at younger age was likely associated with the IGF1 genotype variant not carrying the (CA)19 repeat allele, menarche before the age of 12 and being a current cigarette smoker. Subjects lacking the (CA)19 repeat allele were likely to be at a 1.7 times higher risk at menopause than those homozygous for the (CA)19 repeat allele. The homozygous 19/19 versus heterozygous 19/non-19 difference in median age at menopause accounted for 2.5 years of delay in menopause in homozygous carriers of the (CA)19 repeat allele (52.5 versus 50.0 years).
Significant association between IGF1 gene and timing of natural menopause has been reported by He and colleagues on the basis of large scale GWAS studies.18 Other studies, recently reviewed by He and Murabito, have suggested that other loci may also contribute to the variation in age at natural menopause.37 This is likely that all 17 genetic loci that have been identified so far, might be attributed for variation in menopausal age. It has been presumed that the variation in age at natural menopause, partially under strong genetic control, is a phenotypic trait determined by many genes with additive effects and modulated by widespread environmental exposures over the individual's lifespan.
The CA polymorphism within the IGF1 gene is suggested to be a biological active polymorphism regulating activity of circulating IGF-1. Based on genotype classification and using the most common (CA)19 repeat allele, several studies have reported the association between these genotype variants and circulating IGF-1 levels. However, data in literature are inconsistent. In the first study by Rosen and colleagues, the homozygous 19/19 genotype variant was associated with significantly lower levels of circulating IGF-1.38 Subsequent studies have shown an association of the 19/19 homozygotes with increased circulating IGF-1 levels35,39 or have found no association at all.34,40 Furthermore, significant associations have been established between IGF1 and GH/IGF-1 dependent conditions, i.e. osteoporosis and breast cancer, but here again data are inconsistent.39,41
In addition to the genetic factor, our study revealed that age at menarche and smoking status might be predictors of menopause timing. These results accord with our earlier observations in a population-based sample which showed that younger age at menarche, a normally short menstrual cycle, cigarette smoking, low educational status and a negative health perception are associated with menopause at younger age, while prior use of oral contraceptives and parity are associated with menopause at older age.4 They are also consistent with those of other studies which have reported age at menarche to be directly or inversely associated with timing of menopause.42 This relationship is consistent with the hypothesis that the onset of menopause might be attributed to the loss of ovarian follicles. Thus, it can be suggested that the fewer oocytes a woman has or the sooner and faster they are depleted, the sooner she will experience menopause.
Smoking is the factor most consistently reported to influence the timing of menopause.4,13,14,43 Data in the literature have demonstrated an earlier median or mean age of menopause among smokers; for current smokers versus noncurrent smokers, the difference ranged from 0.8 to 1.7 years. Some researchers suggest that smoking may have an effect on how a woman's body makes or gets rid of estrogen. Alternatively, some researchers believe certain components of cigarette smoke might destroy eggs and accelerate menopause.43
Several limitations of this study have to be kept in mind when interpreting the results. The cross-sectional study design makes it difficult to define risk factors prospectively but we have estimated the association between the IGF1 promoter CA repeat polymorphism and age at natural menopause after controlling for other potential covariates. Furthermore, we cannot fully exclude selection bias in our study as women voluntarily participated in the health programme. However, we believe that the multivariate approach and integration of multiple factors hypothesized to be associated with the natural menopause timing appreciate the strength of this study.
Conclusions
This study provides evidence that a common cytosine-adenine (CA) microsatellite repeat polymorphism in the P1 regulatory region of the IGF1 gene is an independent predictive factor for age at natural menopause also after adjusting for other exposure variables in multivariate modelling. The increased risk of the onset of natural menopause is associated with the absence of (CA)19, early menarche (prior to 12 years of age) and cigarette smoking. Further studies are needed to clarify the biological mechanism of the IGF-1 pathway in the onset of natural menopause.
Acknowledgements
This work was supported by a research grant N N303 109234 of the Polish Ministry of Science and Higher Education.
Ethical approval
This research project was reviewed and approved by the Bioethics Committee in the Poznań University of Medical Sciences, Poznań, Poland. The study participants gave us their written informed consent.
Author contribution
MK led the research project, conceived and designed this paper, performed statistical analyses and wrote first version of the manuscript; AG-J designed and undertook interpretation of molecular genetic data; JP-B carried out molecular genetic analyses; WK, JK and BH-N conducted medical examination, collected data from the interview, and interpreted endocrine test results. All the authors critically read and approved the final version of this manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sievert Leidy L. Menopause; A Biocultural Perspective. New Brunswick, NJ: Rutgers University Press. 2006
2. Gosden RG. Biology of Menopause: The Causes and Consequences of Ovarian Aging. New York: Academic Press. 1985
3. Thomas F, Renaud F, Benefice E. et al. International variability of ages at menarche and menopause: patterns and main determinants. Hum Biol. 2001;73(2):271-290
4. Kaczmarek M. The timing of natural menopause in Poland and associated factors. Maturitas. 2007;57:139-153
5. Dratva J, Gómez Real F, Schindler C. et al. Is age at menopause increasing across Europe? Results on age at menopause and determinants from two population-based studies. Menopause. 2009;16(2):385-394
6. Murray A, Bennett CE, Perry JRB. et al. Common genetic variants are significant risk factors for early menopause: results from the Breakthrough Generations Study. Hum Mol Genet. 2011;20(1):186-192
7. Wellons M, Ouyang P, Schreiner PJ. et al. Early menopause predicts future coronary heart disease and stroke: the Multi-Ethnic Study of Atherosclerosis. Menopause (New York, N.Y.). 2012;19(10):1081-1087
8. Collaborative Group on Hormonal Factors in Breast Cancer. Menarche, menopause, and breast cancer risk: individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol. 2012;13:1141-1151
9. Dossus L, Allen N, Kaaks R. et al. Reproductive risk factors and endometrial cancer: the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2010;127:442-51
10. van Asselt KM, Kok HS, Pearson PL. et al. Heritability of menopausal age in mothers and daughters. Fertil Steril. 2004;82(5):1348-1351
11. Murabito JM, Yang Q, Fox C. et al. Heritability of age at natural menopause in the Framingham Heart Study. J Clin Endocr Metab. 2005;90(6):3427-3430
12. Morris DH, Jones ME, Schoemaker MJ. et al. Familial concordance for age at natural menopause: results from the Breakthrough Generations Study. Menopause. 2011;18(9):956-961
13. Bernis C, Reher DS. Environmental contexts of menopause in Spain: comparative results from recent research. Menopause. 2007;14(4):777-787
14. Gold E, Crawford SL, Avis NE. et al. Factors related to age at natural menopause: longitudinal analyses from SWAN. Am J Epidemiol. 2013;178(1):70-83
15. Stolk L, Zhai G, van Meurs JBJ. et al. Loci at chromosomes 13, 19 and 20 influence age at natural menopause. Nat Genet. 2009;41(6):645-647
16. Stolk L, Perry JRB, Chasman DI. et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44:260-268
17. He Ch, Murabito JM. Genome-wide association studies of age at menarche and age at natural menopause. Mol Cell Endocrinol. 2014;382:767-779
18. He C, Kraft P, Chasman DI. et al. A large-scale candidate-gene association study of age at menarche and age at natural menopause. Hum Genet. 2010;128(5):515-527
19. Hoppener JW, de Pagter-Holthuizen P, Geurts van Kessel AH. et al. The human gene encoding insulin-like growth factor I is located on chromosome 12. Hum Genet. 1985;69:157-160
20. Rotwein P, Pollock KM, Didier DK, Krivi GG. Organization and sequence of the human insulin-like growth factor I gene. Alternative RNA processing produces two insulin-like growth factor I precursor peptides. J Biol Chem. 1986;261:4828-4832
21. Wang L, Wang X, Adamo ML. Two putative GATA motifs in the proximal exon 1 promoter of rat insulin-like growth factor I gene regulate basal promoter activity. Endocrinology. 2000;141:1118-1126
22. Kirkpatrick BW. Identification of a conserved microsatellite site in the porcine and bovine insulin-like growth factor-I gene 5¢ flank. Anim Genet. 1992;23:543-548
23. Mittanck DW, Kim S-W, Rotwein P. Essential promoter elements are located within the 5' untranslated region of human insulin-like growth factor - I exon I. Mol Cell Endocrinol. 1997;126:153-163
24. Fletcher O, Gibson L, Johnson N. et al. Polymorphisms and circulating levels in the insulin-like growth factor system and risk of breast cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:2-19
25. Wagner K, Hemminki K, Forsti A. The GH1/IGF-1 axis polymorphisms and their impact on breast cancer development. Breast Cancer Res Treat. 2007;104:233-248
26. Rosen CJ, Kurland ES, Vereault D. et al. Association between serum insulin growth factor-I (IGF-I) and a simple sequence repeat in IGF-I gene: implications for genetic studies of bone mineral density. J Clin Endocrinol Metab. 1998;83:2286-2290
27. Kato I, Eastham J, Li B. et al. Genotype-phenotype analysis for the polymorphic CA repeat in the insulin-like growth factor-I (IGF-I) gene. Eur J Epidemiol. 2003;18(3):203-209
28. Terry KL, Tworoger SS, Gates MA. et al. Common genetic variation in IGF1, IGFBP1 and IGFBP3 and ovarian cancer risk. Carcinogenesis. 2009;30(12):2042-2046
29. Baker J, Hardy MP, Zhou J. et al. Effects of an Igf1 gene null mutation on mouse reproduction. Mol Endocrinol. 1996;10:903-918
30. Słuczanowska-Głąbowska S, Laszczyńska M, Piotrowska K. et al. Morphology of ovaries in laron dwarf mice, with low circulating plasma levels of insulin-like growth factor-1 (IGF-1), and in bovine GH-transgenic mice, with high circulating plasma levels of IGF-1. J Ovar Res. 2012;5(18):1-8
31. Kaczmarek M. A Polish version of Menopause-Specific Questionnaire, its validity and reliability. Manuscript. 2000
32. Report of a WHO Scientific Group. Research on the menopause. WHO Technical Report Series 670, Geneva: World Health Organisation. 1981
33. van Turenhout EC, Deijen JB, Koppes LLJ. et al. Age- and sex-related interactions between insulin-like growth factor-I promoter polymorphisms and IGF-I levels. The Open Neuroendocrinol J. 2011;4:15-21
34. Jernstrom H, Deal C, Wilkin F. et al. Genetic and nongenetic factors associated with variation of plasma levels of insulin-like growth factor-I and insulin-like growth factor-binding protein-3 in healthy premenopausal women. Cancer Epidem Biomar. 2001;10:377-384
35. Vaessen N, Heutink P, Janssen JA. et al. A polymorphism in the gene for IGF-I: functional properties and risk for type 2 diabetes and myocardial infarction. Diabetes. 2001;50(3):637-642
36. DeLellis K, Ingles S, Kolonel L. et al. IGF1 genotype, mean plasma level and breast cancer risk in the Hawaii/Los Angeles multiethnic cohort. Br J Cancer. 2003;88:277-282
37. He Ch, Murabito JM. Genome-wide association studies of age at menarche and age at natural menopause. Mol Cel Endocrinol. 2014;382:767-779
38. Rosen CJ, Kurland ES, Vereault D. et al. Association between serum insulin growth factor-I (IGF-I) and a simple sequence repeat in IGF-I gene: implications for genetic studies of bone mineral density. J Clin Endocrinol Metab. 1998;83:2286-2290
39. Missmer SA, Haiman CA, Hunter DJ. et al. A sequence repeat in the insulin-like growth factor-1 gene and risk of breast cancer. Int J Cancer. 2002;100:332-336
40. Giovannucci E, Haiman CA, Platz EA. et al. Dinucleotide repeat in the insulin-like growth factor-I gene is not related to risk of colorectal adenoma. Cancer Epidem Biomar. 2002;11:1509-1510
41. Perrini S, Laviola L, Carreira MC. et al. The GH/IGF1 axis and signaling pathways in the muscle and bone: mechanisms underlying age-related skeletal muscle wasting and osteoporosis. J Endocriol. 2010;205:201-210
42. Forman M, Mangini LD, Thelus-Jean R. et al. Life-course origins of the ages at menarche and menopause. Adolescent Health, Medicine and Therapeutics. 2013;4:1-21
43. Sun L, Tan L, Yang F. et al. Meta-analysis suggests that smoking is associated with an increased risk of early natural menopause. Menopause. 2012;19(2):126-132
Author contact
![]() Corresponding author: Maria Kaczmarek, Department of Human Biological Development, Institute of Anthropology, Faculty of Biology, Adam Mickiewicz University in Poznań, Umultowska 89, 61-614 Poznań, Poland. Phone: +48618295759; Fax: +48618295730; E-mail: makacedu.pl
Corresponding author: Maria Kaczmarek, Department of Human Biological Development, Institute of Anthropology, Faculty of Biology, Adam Mickiewicz University in Poznań, Umultowska 89, 61-614 Poznań, Poland. Phone: +48618295759; Fax: +48618295730; E-mail: makacedu.pl

 Global reach, higher impact
Global reach, higher impact