3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2015; 12(1):17-22. doi:10.7150/ijms.10144 This issue Cite
Research Paper
Quantitative Detection of Circulating Nucleophosmin Mutations DNA in the Plasma of Patients with Acute Myeloid Leukemia
1. Key Laboratory of Laboratory Medical Diagnostics Designated by the Ministry of Education, College of Laboratory Medicine, Chongqing Medical University, Yixueyuan Road, Chongqing 400016, P.R.China.
2. Department of Laboratory Medicine, Yantai Yuhuangding Hospital, Yantai 264000, P.R.China.
3. Center for Hematology, Southwest Hospital, Third Military Medical University, Chongqing 400038, P.R.China.
4. Department of Laboratory Medicine, the First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, P.R.China.
Received 2014-7-19; Accepted 2014-10-21; Published 2015-1-1
Abstract
Objective: The aim of this study was to quantify the copies of circulating nucleophosmin (NPM) mutations DNA in the plasma of patients with acute myeloid leukemia (AML) and to explore the association of circulating NPM mutation levels with clinical characteristics.
Design and Methods: The presence of NPM mutations in 100 Chinese patients newly diagnosed with AML were identified by RT-PCR and sequencing analysis. Copies of circulating NPM mutation A (NPM mut.A) DNA in the plasma of mutation-positive cases were quantified by real-time quantitative PCR (qRT-PCR). Furthermore, the association of circulating NPM mutation levels and clinical characteristics was analyzed.
Results: NPM mutations were identified in 37 of the 100 patients and all cases were NPM mut.A. The circulating NPM mut.A levels ranged from 0.35×108 copies/ml to 6.0×108 copies/ml in the 37 mutation-positive cases. The medium and quartile M (P25, P75) of the circulating NPM mut.A levels in patients classified as M2, M4 and M5 morphological subtypes were 1.35×108 (0.76×108, 1.91×108) copies/ml, 1.81×108 (1.47×108, 2.2×108) copies/ml and 2.50×108 (2.42×108, 3.05×108) copies/ml, respectively. Circulating NPM mut.A levels were significantly higher in patients with the M5 subtype of AML compared to patients with the M2 and M4 subtypes (p=0.000, p=0.046). In addition, circulating NPM mut.A copies were significantly associated with a higher white blood cell count, platelet count and bone marrow blast percentage (p<0.05).
Conclusion: Our results suggest that circulating NPM mutations DNA assay serves as a complementary to the routine investigative protocol of NPM-mutated leukemia.
Keywords: nucleophosmin, mutation, acute myeloid leukemia, circulating DNA, real-time quantitative polymerase chain reaction.
Introduction
Circulating DNA in serum or plasma bears the same genetic and epigenetic changes as in a patient's tumor tissues, indicating the possibility of creating less-invasive diagnostic tests based on tumor-specific DNA markers [1]. A number of investigations has revealed circulating DNA to be present in higher levels in patients with cancer of the central nervous system, breast, ovarian, colon, liver, lung, prostate, pancreas and thyroid [2,3]. Plasma samples in patients with leukemia enrich tumor-specific DNA more than in solid tumors because leukocytes exist in the peripheral blood (PB) [4]. The levels of circulating DNA in patients with hematological malignancies, such as chronic myeloid leukemia [5], multiple myeloma [6] and myelodysplastic syndrome [7] are higher than those in healthy individuals. Our previous work showed that the concentration and integrity of circulating DNA in the plasma were significantly increased in patients with acute myeloid leukemia (AML) [8]. However, the leukemia-specific genetic markers in the plasma of patients with AML have not been investigated further.
Nucleophosmin (NPM) is the most commonly mutated gene in AML [9]. NPM mutations destroy the nucleolar localization signal of NPM and cause an abnormal cytoplasmic accumulation of it (NPMc+) [10], which was included as a provisional entity in the revised 2008 World Health Organization (WHO) classification for AML [11]. Over 50 molecular NPM mutation variants have been identified and the most common mutation is NPM mutation A (NPM mut.A) [12]. NPM mutations are fairly stable over the course of the disease [13]. The expression of NPM mutations is associated with therapy response and are strongly predictive for impending hematological relapse [14,15]. Furthermore, the level of NPM mutations is useful for the assessment of minimal residual disease (MRD) [16]. However, the majority of researches has focused on the detection of cellular NPM mutation in bone marrow (BM) samples.
In the present study, NPM mutations were identified from 100 patients with AML, and the number of copies of circulating NPM mutations in these positive cases were determined by quantitative real-time PCR (qRT-PCR). In addition, the association of circulating NPM mutation levels with clinical characteristics was analyzed.
Materials and Methods
Study subjects
A total of 100 primary AML diagnosed at the Southwest Hospital of the Third Military Medical University and the First Affiliated Hospital of Chongqing Medical University in China were studied. All cases were diagnosed through cytomorphology, cytogenetic, and molecular genetic analyses of BM aspirates. All patients were classified according to the French-American-British (FAB) criteria and the cases with FAB subtype M3 were not included in the trial. PB and BM specimens were collected prior to any definitive therapy. This study was conducted in a blind manner. This study has been approved by the Institutional Ethics Review Board for human studies, and all patients signed an informed consent.
Sample collection and circulating DNA isolation
Peripheral blood samples were collected into EDTA-containing vacuum tubes and processed within 24 hours. The plasma and cells samples was separated by centrifugation (3,000 g for 20 min) and stored at -80°C for analysis. Circulating DNA was isolated from 2 ml of the plasma samples using a QIAamp DNA Blood Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
RT-PCR for NPM and sequencing for NPM mutations
The RT-PCR amplification of the NPM gene was performed in a reaction volume of 20 μl, containing 2 μl plasma DNA; according to the manual, 20 pmol of each primer (TaKaRa, Tokyo, Japan), MgCl2, dNTPs, Taq 1× Buffer and DNA Taq polymerase were used. The forward and reverse primers for NPM were: 5'-TTAACTCTCTGGTGGTAGAATGAA-3' and 5'-AAGTTCTCACTCTGCATTATAAAAAGGA-3', respectively. PCR amplification included denaturation at 94°C for 5 min, followed by 35 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 45 sec, with a final elongation at 72°C for 10 min. The amplification products were purified using the Cycle Pure Kit (Omega, Georgia, USA) and were sequenced to verify the NPM gene mutations. To validate the NPM mutation type, PCR products from the mutation-positive samples were T-A cloned into the pMD18-T vector (TaKaRa, Tokyo, Japan) and then transformed into Escherichia coli DH5α. The recombinant plasmid DNA was confirmed by sequencing for the NPM mutation types.
qRT-PCR for circulating NPM mut.A levels
The plasmid vector pEGFPC1-NPM1 mA was kindly provided by Dr. B Falini (Institute of Hematology, University of Perugia, Perugia, Italy). The plasmid DNA concentration was determined by absorbance measurement, and 10-fold serial plasmid dilutions (101 to 1012 copies/ml) were prepared. Sequential dilutions were amplified by qRT-PCR to construct a standard curve for the absolute quantitative assessment of copy number. Each dilution was tested a minimum of 5 times, and standard curves were obtained by plotting the CT value versus the plasmid copy number logarithm. Then, the correlation coefficient of the standard curve was calculated. The reproducibility of this approach was reflected by the coefficients of variation (CV). For measurement of within-run variation, seven different levels of the plasmid were measured 10 times. Each measurement was performed on different days when the between-run variation was measured.
The circulating NPM mut.A levels were determined by qRT-PCR. The 20 μl reaction mixture contained 2 μl of DNA, 1 μl (0.4 μM) of sequence specific primers and 10 μl of SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan). The sequences of the NPM mut.A primers were as follows: forward 5'-AGGCTATTCAAGATCTCTGTCTGG-3', and reverse 5'-AAGTTCTCACTCTGCATTATAAAAAGGA-3'. qRT-PCR was performed on a Rotor-Gene 6000 Real-Time PCR instrument (Corbett Research, Sydney, Australia). The copies of NPM mut.A in each sample were determined using the standard curve. The mean NPM mut.A copy number was calculated from triplicate reactions. All qRT-PCR assays were performed in a blinded fashion, without knowledge of specimen identity.
Statistical analysis
The data were analyzed using the t-test and Spearman correlation test. The circulating NPM mut.A levels were summarized as medium and quartile M (P25, P75). SPSS (Version 16.0) was used for statistical analysis. A value of p<0.05 was considered statistically significant. All clinical data were collected from patient records and organized in the leukemia clinical database.
Results
Clinical information on subjects
One hundred newly diagnosed patients with AML were recruited into the present study (Table 1). By FAB classification, 36 patients were M2, 20 were M4, 23 were M5 and 21 were other subtypes. The relevant clinical characteristics of the AML patients are summarized in Table 1.
Identification of NPM mutations in AML patients
We identified the NPM mutation in 37 (24 females and 13 males) of the 100 samples from the AML patients (Table 2) and all cases were NPM mut.A, with a duplication of the TCTG tetranucleotide at position 956-959 of the reference sequence (NM_002520). As shown in Table 2, NPM mutations were more frequent in the patients classified as M4 (9/20) and M5 (14/23) subtypes when compared with other subtypes, while the NPM mutations were not detected in patients with the M0 (0/2) and M6 (0/4) subtypes. In addition, among the 37 patients positive for NPM mutations, 35 harbored NPM mutations in the detection of PB mononuclear cells.
Clinical characteristics of AML patients.
| Total cohort, n=100 | |||
| Sex, Male / Female | 56 / 44 | ||
| Median age, y (range) | 37 (18-62) | ||
| Median WBC count, ×109/L (range) | 17.5 (0.7-280.0) | ||
| Median platelet count, ×109/L (range) | 61.4 (10.0-655.0) | ||
| FAB classification | |||
| M0 | n=2 | 2.0% | |
| M1 | n=15 | 15.0% | |
| M2 | n=36 | 36.0% | |
| M4 | n=20 | 20.0% | |
| M5 | n=23 | 23.0% | |
| M6 | n=4 | 4.0% | |
WBC: white blood cell; FAB classification: French-American-British classification, a classification of acute leukemia produced by three-nation joint collaboration.
The positive cases of NPM mutations in AML patients.
| AML classification | Number of cases | Number of positive cases (%) | |
|---|---|---|---|
| Plasma | PB cells | ||
| M0 | 2 | 0 (0) | 0 (0) |
| M1 | 15 | 2 (13) | 2 (13) |
| M2 | 36 | 12 (33) | 11 (31) |
| M4 | 20 | 9 (45) | 8 (40) |
| M5 | 23 | 14 (61) | 14 (61) |
| M6 | 4 | 0 (0) | 0 (0) |
Methodological evaluation of qRT-PCR for NPM mut.A
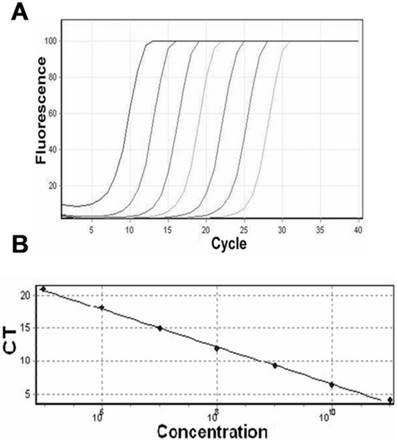
The plasmid carrying target NPM mut.A gene was successfully constructed. As shown in Fig. 1A, every dilution (105 ~1011 copies/ml) of the target plasmid showed typical amplification curves. The corresponding qRT-PCR standard curve showed an ideal linear correlation between the CT values and the logarithm of the initial template concentration, with a correlation coefficient of 0.99812 (Fig. 1B). Additionally, the repeatability of the qRT-PCR assays was estimated. The within-run and between-run CV of this qRT-PCR were 3.21% (n=10) and 4.87% (n=10), respectively.
Circulating NPM mut.A DNA in AML patients
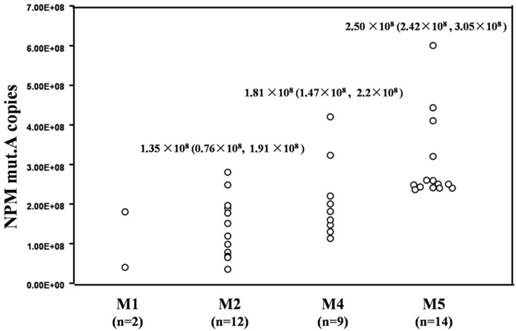
Circulating NPM mut.A DNA levels from the 37 NPM mutation-positive cases were determined by qRT-PCR. The NPM mut.A copy numbers ranged from 0.35×108 copies/ml to 6.0×108 copies/ml. The medium and quartile M (P25, P75) of the NPM mut.A copy numbers in the cases classified as M2, M4 and M5 subtypes were 1.35×108 (0.76×108, 1.91×108) copies/ml, 1.81×108 (1.47×108, 2.2×108) copies/ml and 2.50×108 (2.42×108, 3.05×108) copies/ml, respectively. Circulating NPM mut.A levels were significantly higher in patients with the M5 subtype compared to patients with the M1, M2 and M4 subtypes (p=0.032, p=0.000, p=0.046), and there was no statistically significant differences in the circulating NPM mut.A levels between patients with the other subtypes (p>0.05). As shown in Fig. 2.
Establishment of the standard curve for qRT-PCR. (A) Representative amplification plots of serial plasmid dilutions ranging from 105 to 1011 copies/ml in each reaction. (B) Standard curve of the real-time amplification of NPM mut.A derived from plots in (A) with a correlation of 0.99812.
Circulating NPM mut.A DNA and clinical characteristics
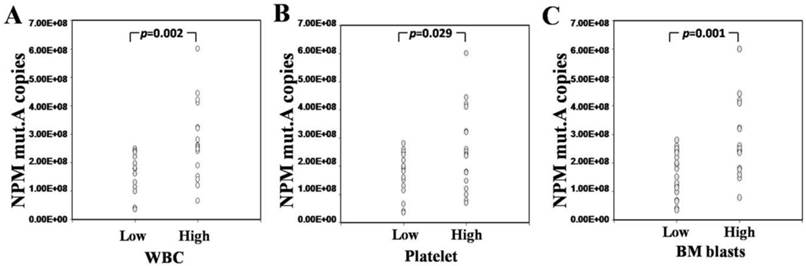
To investigate the correlations between circulating NPM mut.A DNA and clinical characteristics, we reviewed the participants' white blood cell (WBC) counts, platelet counts, hemoglobin levels and BM blast percentages. All the laboratory parameters were categorized into two groups (low/high). The circulating NPM mut.A DNA was significantly higher in the patients with a high peripheral WBC count (>40×109 /L), high platelet count (>70×109 /L) and high BM blast percentage (>70%), compared to the patients with low values for those laboratory parameters (p=0.002; p =0.029; p=0.001), as shown in Fig. 3. No significant correlation between circulating NPM mut.A level and hemoglobin level was found (p>0.05, data not shown). In addition, a higher NPM mut.A level was frequently found in elder patients (>60 years); there was no significant difference in NPM mut.A level between males and females (p>0.05).
Discussion
An increasing number of reports have demonstrated the molecular diagnostic value of PB circulating DNA in patients with hematological malignancies. Circulating DNA of leukemia patients is currently available but is nonspecific and has a low sensitivity. NPM gene mutations are the most common genetic alteration in AML, and the NPM mutation copies are clinically important for monitoring MRD [17,18]. We detected the circulating NPM mut.A DNA levels and found the clinical role of circulating NPM mut.A DNA in AML.
Circulating NPM mut.A copies in patients with AML. The circulating NPM mut.A copy numbers in 37 NPM mutation-positive cases were determined by qRT-PCR assay and summarized as median and quartile M (P25, P75).
Circulating NPM mut.A copies and laboratory parameters of NPM-mutated AML. Dot plots show the distribution of NPM mut.A copies in AML patients. Each laboratory parameter (WBC count, platelet count and BM blast percentage) was divided into two categories (low/high).
We identified NPM mutations from 100 patients with AML. Thirty-seven NPM mutation-positive cases were identified. A high NPM mutation rate was found in the patients with M4 (9/20) and M5 (14/23) subtypes, which was in accordance with the results of Falini B et al [13]. The NPM mutation was absent in patients with the M0 (0/2) and M6 (0/4) subtypes, which should be confirmed in larger studies. In the present study, we compared the NPM mutations between the plasma and PB cells. Of interest, we identified 37 of 100 AML plasma samples positive for NPM mutations, while only 35 patients identified in PB cell samples. This discrepancy was also observed in the study of Ma W et al [19] in which they identified NPM mutations in 24 of 98 AML plasma samples, while cell samples showed positivity in only 22 patients.
To quantitatively detect the circulating NPM gene mutation DNA copies, we first established the qRT-PCR assays for the detection of circulating NPM mut.A gene copies. High correlation coefficients (0.99812) allowed accurate assessment of the quantity of NPM mut.A. Good reproducibility (within-run CV of 3.21% and between-run CV of 4.87%) showed that the established qRT-PCR protocol in our study was effective. Then, circulating NPM mut.A DNA was detected and ranged from 0.35×108 copies/ml to 6.0×108 copies/ml in the 37 mutation-positive cases, and higher circulating NPM mut.A levels were found in the patients with the M5 subtype compared to the patients with M2 and M4 subtypes (p=0.000, p=0.046). Indeed, Falini B et al [20] reported that NPM gene mutations occur more frequently in M5 and M4. Furthermore, we observed the clinical relevance of circulating NPM mutations with clinical parameters according to the study of Döhner K et al [21]. We found that the circulating NPM mut.A copies in AML patients were associated with higher WBC count, higher platelet count and higher BM blast percentage. This is in agreement with previous study by Ehninger G et al [22], which indicated that the presence of NPM mutations was associated with higher WBC count, higher platelet count and higher BM blast percentage.
In conclusion, we quantitatively detected circulating NPM mut.A gene copies by an qRT-PCR method that we established and found that elevated NPM mut.A gene copies were associated with clinical characteristics. Our data support the utility of circulating NPM mut.A DNA serves as a complementary to the routine investigative protocol of NPM-mutated leukemia. It is important to note that the sample size of our study was small, and prospective studies in large patient series are needed to evaluate further the clinical relevance of circulating NPM mut.A for therapy response and clinical outcomes.
Abbreviations
NPM: nucleophosmin; AML: acute myeloid leukemia; qRT-PCR: real-time quantitative polymerase chain reaction; WHO: World Health Organization; FAB: French-American-British; PB: peripheral blood; BM: bone marrow; WBC: white blood cell; MRD: minimal residual disease.
Acknowledgements
This project was funded by the National Natural Science Foundation of China (No. 81271913) and by the Chongqing Natural Science Foundation of CQ CSTC (No. csct2013jcyjA10035).
Competing interests
The authors have declared that no competing interest exists.
References
1. Vlassov VV, Laktionov PP, Rykova EY. Circulating nucleic acids as a potential source for cancer biomarkers. Curr Mol Med. 2010;10:142-65
2. Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta. 2007;1775:181-232
3. Elshimali YI, Khaddour H, Vadgama JV. et al. The Clinical Utilization of Circulating Cell Free DNA (CCFDNA) in Blood of Cancer Patients. Int J Mol Sci. 2013;14:18925-58
4. Giles FJ, Albitar M. Plasma-based testing as a new paradigm for clinical testing in hematologic diseases. Expert Rev Mol Diagn. 2007;7:615-23
5. Ma W, Tseng R, Albitar M. et al. Plasma RNA as an alternative to cells for monitoring molecular response in patients with chronic myeloid leukemia. Haematologica. 2007;92:170-5
6. Dalay N, Gezer U, Holdenrieder S. et al. Correlation of histone methyl marks with circulating nucleosomes in blood plasma of cancer patients. Oncol Lett. 2012;3:1095-8
7. Gezer U, Mert U, Naoe T. et al. Using peripheral blood circulating DNAs to detect CpG global methylation status and genetic mutations in patients with myelodysplastic syndrome. Biochem Biophys Res Commun. 2012;419:662-9
8. Gao YJ, He YJ, Zhang L. et al. Increased integrity of circulating cell-free DNA in plasma of patients with acute leukemia. Clin Chem Lab Med. 2010;48:1651-6
9. Falini B, Mecucci C, Martelli MF. et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254-66
10. Falini B, Bolli N, Nicoletti I. et al. Both carboxyterminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514-23
11. H.J.M. de Jonge, G. Huls, E.S.J.M. de Bont. Gene expression profiling in acute myeloid leukaemia. Neth J Med. 2011;69:167-76
12. Rau R, Brown P. Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity. Hematol Oncol. 2009;27:171-81
13. Meloni G, Mancini M, Falini B. et al. Late relapse of acute myeloid leukemia with mutated NPM after eight years: evidence of NPM mutation stability. Haematologica. 2009;94:298-300
14. Kristensen T, Møller MB, Preiss B. et al. NPM1 mutation is a stable marker for minimal residual disease monitoring in acute myeloid leukaemia patients with increased sensitivity compared to WT1 expression. Eur J Haematol. 2011;87:400-8
15. Bacher U, Badbaran A, Kröger N. et al. Quantitative monitoring of NPM1 mutations provides a valid minimal residual disease parameter following allogeneic stem cell transplantation. Exp Hematol. 2009;37:135-42
16. Dvorakova D, Racil Z, Mayer J. et al. Monitoring of minimal residual disease in acute myeloid leukemia with frequent and rare patient-specific NPM1 mutations. Am J Hematol. 2010;85:926-9
17. Krönke J, Schlenk RF, Döhner K. et al. Monitoring of minimal residual disease in NPM-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol. 2011;29:2709-16
18. Schnittger S, Kern W, Haferlach T. et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114:2220-31
19. Ma W, Kantarjian H, Zhang X, Jilani I. et al. Detection of nucleophosmin gene mutations in plasma from patients with acute myeloid leukemia: clinical significance and implications. Cancer Biomark. 2009;5:51-8
20. Falini B, Gionfriddo I, Cecchetti F. et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): any hope for a targeted therapy? Blood Rev. 2011;25:247-54
21. Döhner K, Schlenk RF, Habdank M. et al. Mutant nuclephosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740-6
22. Thiede C, Koch S, Ehninger G. et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006;107:4011-20
Author contact
![]() Corresponding author: Ling Zhang, College of Laboratory Medicine, Chongqing Medical University, No.1, Yixueyuan Road, Chongqing, 400016, China. Tel: +86 023-68485240 Fax: +86 023-68485239 Email: lingzhangedu.cn.
Corresponding author: Ling Zhang, College of Laboratory Medicine, Chongqing Medical University, No.1, Yixueyuan Road, Chongqing, 400016, China. Tel: +86 023-68485240 Fax: +86 023-68485239 Email: lingzhangedu.cn.

 Global reach, higher impact
Global reach, higher impact