Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(12):1270-1274. doi:10.7150/ijms.9343 This issue Cite
Research Paper
A Novel PCR-Based Approach to Discover miRNA Target Genes
1. Zhejiang Provincial Key Laboratory of Pathophysiology, School of Medicine, Ningbo University, Ningbo, ZJ 315000, China
2. The Neurology Department of the 148th Hospital of PLA, Zibo, SD 255300, China
3. Department of Medicine, University of Chicago, Chicago, IL 60637, USA
4. Biological Science Division, University of Chicago, Chicago, IL 60637, USA
5. Department of Psychiatry, University of Chicago, Chicago, IL 60637, USA.
*SD and YW contributed equally to the work.
Received 2014-4-9; Accepted 2014-9-9; Published 2014-10-11
Abstract
MiRNAs are potent regulators of gene expression, and most miRNAs have from several to several thousands of gene targets. Validating the numerous gene targets of a given miRNA remains challenging despite the existence of various tools and databases that predict candidate gene-miRNA pairs. In the present study, we present a high-throughput but flexible method that applies a PCR-based application to simulate the binding of miRNAs to their gene targets. Using hsa-miR-377 as an illustrative example, our method was able to identify 13 potential targets of hsa-miR-377. Moreover, our results include 2 genes (SOD2 and PPM1A) that have already been verified as targets of hsa-miR-377. Our method may provide an alternative way of identifying the gene targets of miRNAs for future research.
Keywords: miRNA, gene target, hsa-miR-377, high-throughput.
Introduction
Mature miRNAs are a class of small single-stranded RNAs roughly 20 nts long. They are potent regulators of gene expression and are predicted to modulate the expression of several to several thousands of genes. MiRNAs act through pair-matching of their seed sequence to the 3'-untranslated regions of gene transcripts [1]. Identifying the gene targets of miRNAs is crucial for illustrating the biological mechanisms underlying these powerful regulatory molecules. Elucidating the role of miRNA regulation on gene expression has contributed to the mechanistic understanding of cardiogenesis [2], skeletal muscle proliferation and differentiation [3], drug resistance [4], and cancer metastasis [5]. Dysfunction of miRNA-gene targeting may lead to many diseases such as cancer [5], complex psychiatric disorder [6], and addiction [7].
While a number of computational algorithms have been published to predict the target genes of a given miRNA, huge discrepancies exist in the predicted results of these algorithms [8]. Although there are curated databases on miRNA-gene targeting relationships [9] (http://diana.cslab.ece.ntua.gr/tarbase/), the validation of these various bioinformatic predictions remains a challenge for researchers.
In the present studies, we propose a novel PCR-based chip assay that can be used to supplement the existing approaches. It is based on the seed-match theory between miRNA sequences and the 3'-UTR of their target mRNAs [10, 11]. Coupled with gene expression microarray, this PCR-based in-vitro approach can be used to quantify the binding ability of miRNAs to target mRNAs.
Methods and materials
First-strand cDNA synthesis and the 3'-UTR amplification of miR-377 target genes
The total RNAs of normal human mesangial cells were extracted with TRIZOL (Invitrogen Corporation, Carlsbad, CA, USA). Details of cell culture are as described previously [12]. As shown in Figure 1, total RNA was reversely transcribed to cDNA using a P-oligo-dT primer with a 5'-overhanged sequence (5'-GGCTAGTCTCGTGATCGA-3'). Then the cDNA mixture was used for PCR amplication using a tag primer (5'-GGCTAGTCTCGTGATCGA-3') and hsa-miR-377 derived primer (5'-ACAAAAGTTGCCTTTGTGTgAT-3'; the “g“ stands for a lock nucleotide acid) or its control primer with 3 mutations (a, c and t) in the underlined region complemented to hsa-miR-377 (5'-ACAAATGTAGCGTTaGTcTGtT-3'). PCR was performed as follows: 3 min at 95ºC followed by 5 cycles of amplification (30 sec at 95ºC, 30 sec at 45ºC, 2 min at 72 ºC), and another 30 cycles (30 sec at 95ºC, 30 sec at 54ºC, 2 min at 72 ºC) for further amplification.
Quantification of the amplified 3'-UTRs in cDNA pools and identification of target genes of miR-377
PCR products were labeled (Bioprimer DNA labeling System, Invitrogen) and hybridized with human expression microarrays (GeneChip Human Gene 1.0 ST Array; Affymetrix, Santa Clara, CA, USA). The image data were analyzed by DNA-Chip Analyzer (http://www.dchip.org) to generate a normalized signal for each gene's exon on the array. The potential target genes of hsa-miR-377 met the following criteria. Firstly they must have probes in 3'-UTRs with a signal intensity of at least 1000. Secondly, they need to have probes in other regions (coding sequences (CDS) or 5'-UTR) with a signal intensity of less than 200. Finally, the ratio of the signal intensity of 3'-UTRs between miR-377 derived primer PCR input and its control PCR input has to be greater than 10 for these miR-377 target genes. In the current microarray, the signal intensity of 200 suggests a moderate amount of capturing the PCR products using the primer sets. For the same gene, a >1000 value in the 3-UTR and <200 value in other gene regions suggests an enrichment of 3'-UTR targeting in the seed-containing primer set, implying a potential targeting between miR-377 seed sequence and the identified gene.
Results
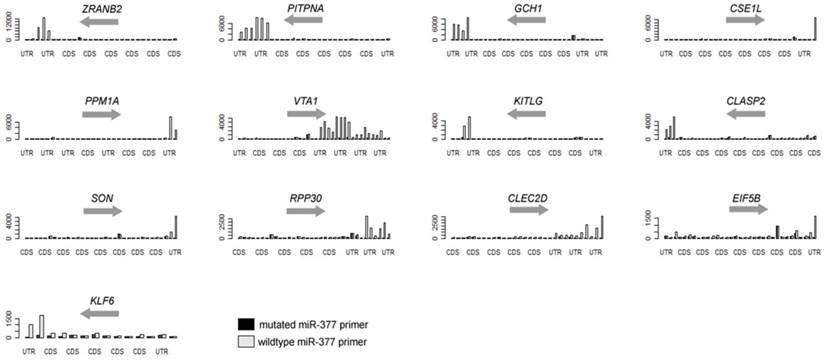
In the present study, we are able to identify 13 genes, as listed in Table 1 and Figure 2. Eight of them (over 60%) are also predicted to miR-377 targets [11, 13-16]. Other genes with less stringent criteria are shown in the Supplementary Material: Table S1. A list of 16,319 transcripts were harvested by sorting the ratio of the 3'-UTR probe signal intensity between miR-377 derived primer PCR input and miR-377 mutated primer PCR input (Supplementary Material: Table S2). The fasta sequences of the 3'-UTRs of transcripts are retrieved from the ENSEMBL website (http://www.ensembl.org/index.html). By using the 8-letter enrichment analysis of Sylmer, we find that “TTGTGTGA” as the seed of miR-377 is significantly enriched in the UTRs of the top transcripts (Figure 1).
As shown in Figure 1, our method comprises of a first strand cDNA synthesis of the transcriptome and a subsequent PCR to amplify the 3'-UTR regions using the miRNA-complement primers. The background noise is controlled by the PCR of another primer with a seed-mutated miRNA sequence coupled with the tag primer. After the PCR procedure, the products are labeled and hybridized onto gene expression array. The levels of the signal-to-noise ratios of the probes in the 3'-UTRs of genes reflect their binding capability to miRNAs.
Using hsa-miR-377 as an example, we coupled the tag primer with the primer complementary to the sequence of hsa-miR-377. In addition, the control PCR uses the tag primer and a primer with mismatches in the seed region of hsa-miR-377. By ranking the signal-to-noise ratio of amplified genes, we find that there is a significant enrichment of miR-377 seed (TTGTGTGA) in the 3'-UTR of the ranked mRNAs using the Sylamer software [17]. Additionally, 8 of the aforementioned 13 genes were previously predicted to be miR-377 targets by various in-silico methods (Table 1 and Supplementary Material: Table S1). SOD2 (Supplementary Material: Table S2) and PPM1A (Table 1) have already been validated as targets of miR-377 in the previous studies [12].
The target genes of hsa-miR-377 a.
| Gene | Entrez id | Ratio | Target predictions |
|---|---|---|---|
| ZRANB2 | 9406 | 305.73 | |
| PITPNA | 5306 | 161.58 | |
| GCH1 | 2643 | 137.16 | PITA |
| CSE1L | 1434 | 123.82 | |
| PPM1A | 5494 | 104.08 | miRanda, PITA |
| VTA1 | 51534 | 98.83 | TargetScan_nonconserved |
| KITLG | 4254 | 96.78 | TargetScan_nonconserved, PITA |
| CLASP2 | 23122 | 87.43 | miRanda, TargetScan_nonconserved, PITA |
| SON | 6651 | 64.23 | miRanda, TargetScan_nonconserved, PITA |
| RPP30 | 10556 | 59.19 | TargetScan_nonconserved, PITA |
| CLEC2D | 29121 | 42.76 | |
| EIF5B | 9669 | 31.75 | miRanda, TargetScan_conserved, PITA |
| KLF6 | 1316 | 23.07 |
a: Ratio is the ratio of the 3'-UTR probe signal intensity between miR-377 derived primer PCR input and miR-377 mutated primer PCR input.
The workflow of the PCR-based approach to identify miRNA target genes.
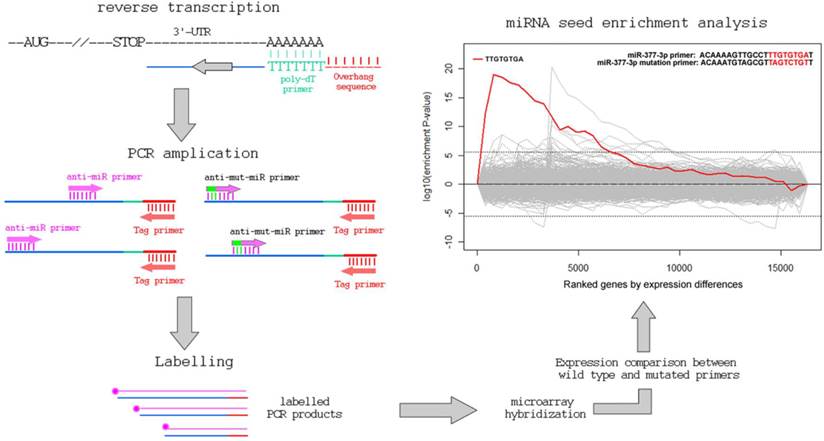
Over-representative 3'-UTRs of the hsa-miR-377 target genes in the PCR.
Discussions
Using hsa-miR-377 as an example, we found 13 genes that potential interact with hsa-miR-377, a miRNA associated with diabetes nephropathy [12] and cancer progression [18]. Among the identified target genes of hsa-miR-377, at least three [GCH1 [19], PPM1A [20-22] and KLF6 [23, 24]] were shown to be linked to the pathogenesis of diabetes and five [GCH1 [25, 26], CSE1L [27], PPM1A [28], KITLG [29] and KLF6 [30]] were shown to be linked to the pathogenesis of cancer. These potential targets of hsa-miR-377 are likely to provide hints for future research on the contribution of hsa-miR-377 in diabetes and cancers.
The classic method for validating the gene targets of a miRNA is the luciferase reporter assay [31], which is designed to test whether or not there is a reduction in luciferase enzyme activity through the miRNA binding to the foreign sequence in the 5'-flanking region of the luciferase gene. Although this method is accurate and thus has become the standard method, it is also labor-intensive and low-throughput. Other high-throughput methods include the anti-miRNA knock down assay [32] and the immunoprecipitation-based chip (ChIP-chip) or sequencing assays (ChIP-Seq) [33]. While the anti-miRNA knock down assay can generate a list of associated genes, it is hard to distinguish between the primary gene targets and other genes that are subsequently influenced by non-miRNA factors. The most recent Argonaute immunoprecipitation-based chip or sequencing arrays have become a very successful approach that can capture the miRNA and its gene targets simultaneously. However, it seems that only the most abundant miRNAs are suitable for this approach.
Our method simulates in vivo binding between a miRNA and its targets. And it can be easily expanded to discover the differences in the gene regulation among the genome-wide miRNAs by comparing the PCR signals from different miRNA primer sets. This method can potentially be used to identify novel miRNA binding targets by employing next generation sequencing techniques to sequence the PCR products. However, there are some limitations to this system. For examples, the oligo-dT primer can't pick up genes lacking 3'-UTR sequences. And for some mature miRNAs with length fewer than 18 bps, this method may fail to design eligible primers due to the severe nonspecific PCR amplification events.
In summary, our study uses a PCR-based approach to simulate the binding between miRNA and the 3'-UTRs of their target genes. This technique is able to reveal novel target genes of a given miRNA in a high-throughput manner. Using human miR-377 as an example, our study shows that the top ranked target genes are significantly enriched with miR-377 seed matched regions in their 3'-UTRs. This approach may provide an alternative way of revealing the targets of miRNAs for future research.
Supplementary Material
Table S1: The list of genes with significant enrichment of the hsa-miR-377 seed-matched sequence in the 3'-UTRs. Table S2: The gene list for Sylmer program.
Abbreviations
miRNAs: MicroRNAs; PCR: Polymerase Chain Reaction; UTR: Untranslated Regions; cDNA: complementary DNA.
Acknowledgements
The research was supported by the grants from the National Natural Science Foundation of China (31100919 and 81371469), Natural Science Foundation of Zhejiang Province (LR13H020003), K.C. Wong Magna Fund in Ningbo University, Ningbo Social Development Research Projects (2012C50032).
Conflict of interest
None of the authors have any commercial or other associations that might pose a conflict of interest. All authors are responsible for the content and writing of the paper.
References
1. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J. et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-73 doi:10.1038/nature03315
2. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214-20 doi:10.1038/nature03817
3. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature genetics. 2006;38:228-33 doi:10.1038/ng1725
4. Beretta GL, Gatti L, Perego P, Zaffaroni N. Camptothecin resistance in cancer: insights into the molecular mechanisms of a DNA-damaging drug. Current medicinal chemistry. 2013;20:1541-65
5. Li M, Fu W, Wo L, Shu X, Liu F, Li C. miR-128 and its target genes in tumorigenesis and metastasis. Experimental cell research. 2013;319:3059-64 doi:10.1016/j.yexcr.2013.07.031
6. O'Connor RM, Dinan TG, Cryan JF. Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Molecular psychiatry. 2012;17:359-76 doi:10.1038/mp.2011.162
7. Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A. et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcoholism, clinical and experimental research. 2010;34:575-87 doi:10.1111/j.1530-0277.2009.01126.x
8. Reyes-Herrera PH, Ficarra E. One decade of development and evolution of microRNA target prediction algorithms. Genomics, proteomics & bioinformatics. 2012;10:254-63 doi:10.1016/j.gpb.2012.10.001
9. Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M. et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic acids research. 2012;40:D222-9 doi:10.1093/nar/gkr1161
10. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33 doi:10.1016/j.cell.2009.01.002
11. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20 doi:10.1016/j.cell.2004.12.035
12. Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X. et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22:4126-35 doi:10.1096/fj.08-112326
13. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92-105 doi:10.1101/gr.082701.108
14. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nature genetics. 2007;39:1278-84 doi:10.1038/ng2135
15. Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91-105 doi:10.1016/j.molcel.2007.06.017
16. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome biology. 2003;5:R1. doi:10.1186/gb-2003-5-1-r1
17. van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nature methods. 2008;5:1023-5 doi:10.1038/nmeth.1267
18. Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro E, Levine AJ. et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2013 doi:10.1038/onc.2013.451
19. Liao YF, Zeng TS, Chen LL, Li YM, Yu F, Hu LJ. et al. Association of a functional polymorphism (C59038T) in GTP cyclohydrolase 1 gene and Type 2 diabetic macrovascular disease in the Chinese population. Journal of diabetes and its complications. 2010;24:313-9 doi:10.1016/j.jdiacomp.2009.04.003
20. Voss M, Paterson J, Kelsall IR, Martin-Granados C, Hastie CJ, Peggie MW. et al. Ppm1E is an in cellulo AMP-activated protein kinase phosphatase. Cellular signalling. 2011;23:114-24 doi:10.1016/j.cellsig.2010.08.010
21. Moll AG, Lindenmeyer MT, Kretzler M, Nelson PJ, Zimmer R, Cohen CD. Transcript-specific expression profiles derived from sequence-based analysis of standard microarrays. PloS one. 2009;4:e4702. doi:10.1371/journal.pone.0004702
22. Lee JE, Lee JS, Hwang SH. Microarray for genes associated with signal transduction in diabetic OLETF keratocytes. Korean journal of ophthalmology: KJO. 2007;21:111-9
23. Qi W, Holian J, Tan CY, Kelly DJ, Chen XM, Pollock CA. The roles of Kruppel-like factor 6 and peroxisome proliferator-activated receptor-gamma in the regulation of macrophage inflammatory protein-3alpha at early onset of diabetes. The international journal of biochemistry & cell biology. 2011;43:383-92 doi:10.1016/j.biocel.2010.11.008
24. Qi W, Chen X, Holian J, Tan CY, Kelly DJ, Pollock CA. Transcription factors Kruppel-like factor 6 and peroxisome proliferator-activated receptor-{gamma} mediate high glucose-induced thioredoxin-interacting protein. The American journal of pathology. 2009;175:1858-67 doi:10.2353/ajpath.2009.090263
25. Pud D, Har-Zahav G, Laitman Y, Rubinek T, Yeheskel A, Ben-Ami S. et al. Association between variants of 5-hydroxytryptamine receptor 3C (HTR3C) and chemotherapy-induced symptoms in women receiving adjuvant treatment for breast cancer. Breast cancer research and treatment. 2014;144:123-31 doi:10.1007/s10549-014-2832-y
26. Pickert G, Lim HY, Weigert A, Haussler A, Myrczek T, Waldner M. et al. Inhibition of GTP cyclohydrolase attenuates tumor growth by reducing angiogenesis and M2-like polarization of tumor associated macrophages. International journal of cancer Journal international du cancer. 2013;132:591-604 doi:10.1002/ijc.27706
27. Tai CJ, Hsu CH, Shen SC, Lee WR, Jiang MC. Cellular apoptosis susceptibility (CSE1L/CAS) protein in cancer metastasis and chemotherapeutic drug-induced apoptosis. Journal of experimental & clinical cancer research: CR. 2010;29:110. doi:10.1186/1756-9966-29-110
28. Lin X, Chen Y, Meng A, Feng X. Termination of TGF-beta superfamily signaling through SMAD dephosphorylation--a functional genomic view. Journal of genetics and genomics = Yi chuan xue bao. 2007;34:1-9 doi:10.1016/S1673-8527(07)60001-0
29. Amagai Y, Tanaka A, Matsuda A, Jung K, Ohmori K, Matsuda H. Stem cell factor contributes to tumorigenesis of mast cells via an autocrine/paracrine mechanism. Journal of leukocyte biology. 2013;93:245-50 doi:10.1189/jlb.0512245
30. Andreoli V, Gehrau RC, Bocco JL. Biology of Kruppel-like factor 6 transcriptional regulator in cell life and death. IUBMB life. 2010;62:896-905 doi:10.1002/iub.396
31. Wang B, Doench JG, Novina CD. Analysis of microRNA effector functions in vitro. Methods. 2007;43:91-104 doi:10.1016/j.ymeth.2007.04.003
32. Kreth S, Ledderose C, Schutz S, Beiras A, Heyn J, Weis F. et al. MicroRNA-150 inhibits expression of adiponectin receptor 2 and is a potential therapeutic target in patients with chronic heart failure. The Journal of heart and lung transplantation: the official publication of the International Society for Heart Transplantation. 2013 doi:10.1016/j.healun.2013.10.014
33. Cullum R, Alder O, Hoodless PA. The next generation: using new sequencing technologies to analyse gene regulation. Respirology. 2011;16:210-22 doi:10.1111/j.1440-1843.2010.01899.x
Author contact
![]() Corresponding author: Qiang Wang (qwanguchicago.edu).
Corresponding author: Qiang Wang (qwanguchicago.edu).

 Global reach, higher impact
Global reach, higher impact