Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(11):1147-1153. doi:10.7150/ijms.9412 This issue Cite
Research Paper
Impact of Single Nucleotide Polymorphism in IL-4, IL-4R Genes and Systemic Concentration of IL-4 on the Incidence of Glioma in Iraqi Patients
1. Medical Research Unit, School of Medicine, University of AL-Nahrain Iraq, Baghdad
2. Biotechnology Department, School of Science, University of AL-Nahrain, Iraq, Baghdad
3. Iraqi National Cancer Center, University of AL-Mustansiriya, Iraq, Baghdad
4. Neuroscience Hospital, Neurosurgery, Iraq, Baghdad
5. Center for Translational Studies, Medical Services, VA Medical Center, Department of Veteran Affairs, Washington DC, USA
6. Department of Pathology, Microbiology and Immunology, School of Medicine, University of South Carolina, SC, USA
Received 2014-4-16; Accepted 2014-8-6; Published 2014-8-22
Abstract
Glioma is the most common and believed to be one of the most aggressive tumors of the central nervous system (CNS) in humans. Very little information is available on the etiology and pathogenesis of these tumors to date. A significant gap remains in our current understanding of the molecular pathways involved in the genesis, progression and clinical behavior of these tumors. Recently, several single nucleotide polymorphisms (SNPs) have been identified in cytokine gene sequences, particularly within the promoter region of these genes, and have been shown to be associated with the development of different types of brain tumors. The present study investigates the association of C-33T SNP in the interleukin-4 (IL-4) gene with systemic IL-4 level and the S503P SNP in the IL-4R gene with the incidence of glioma.
Blood samples were collected from 100 histologically confirmed adult patients with glioma, and 30 apparently healthy individuals from the same area. DNA was extracted from each blood sample, and the IL-4 and IL-4R genes were amplified using polymerase chain reaction (PCR) with gene-specific primers. Systemic IL-4 concentration was assessed in serum samples from each participant by enzyme-linked immunosorbent assay (ELISA). We observed a negative association between the homozygous genotype (CC) of the SNP C-33T of the IL-4 gene with the incidence of glioma (OR=0.19, 95% CI=0.035-1.02), while the T allele of the SNP demonstrated a significant protective association against glioma. Similarly, the heterozygous (CT) and homozygous mutant (CC) of the SNP S503P of the IL-4R gene demonstrated a significant association with glioma development (OR=0.405, 95% CI=0.17-0.969 and OR=0.147, 95% CI=0.036-0.6 respectively), while the C allele exhibited a highly significant association with protection from glioma formation.
These findings suggest that the T allele of the SNP C-33T in the IL-4 gene and the C allele of the SNP S503P in IL-4R may have a protective role against glioma development.
Keywords: Single nucleotide polymorphism (SNPs), interleukin-4, interleukin-4R, brain tumor, and Glioma
Introduction
Tumors of the central nervous system (CNS) represent about 2% of all cancers. Glioma is the most common tumor of the CNS and in adults accounts for almost 80% of primary malignant brain tumors [1]. The most common type of glioma (glioblastoma multiform) is associated with a median survival of only 12-15 months [2]. In Iraq, as published in the cancer registry, CNS tumors are the fifth most common tumor in adults and the second most common in children. However, these tumors are the most challenging regarding their cure due to lots of complications. For example, the total surgical removal of an organ or tumor, which is used with other cancers, cannot be applied to cure brain tumors because each region of the brain has a vital function [3].
Although glioma is considered to have multiple causes, the only established environmental risk factors are ionizing radiation and ultraviolet light [3, 4]. However, this can only explain a small number of gliomas diagnosed as exposure to radiation is relatively rare, and only a minimal percentage of individuals exposed to radiation eventually develop a glioma [5]. Previous studies have reported a link between the immune system and gliomas, and T-lymphocyte infiltrations have been observed in the tumor regions of certain glioma patients [6]. However, elevated numbers of T cells have been correlated with a better survival in patients with Glioblastoma multiform (GBM) [6].
Several studies have examined the effects of single nucleotide polymorphisms (SNPs) in inflammation-associated genes on the risk of developing glioma and focused on their potential role in glioma survival. A previous etiological study has provided evidence that polymorphisms in the IL-4 and IL-4R genes may play an important role in the pathways that regulate glioma development by influencing IgE levels, thus potentially impacting the effectiveness of treatment via more direct cell signaling pathways [7]. However, there are no reports on the association between the polymorphism of cytokine genes and the development of glioma in the Iraqi population.
IL-4 is one of the key players in the immune system, which is secreted from T helper 2 (Th2) cells, NK cells, mast cells, and basophils [8]. IL-4 is a pleiotropic cytokine with multiple functions like Th2 cell development, T and B cell growth factor, and IgE/IgG1 class switching. A large number of single nucleotide polymorphisms (SNPs), have been identified in the IL-4 gene [9], many of which are studied with respect to their association with brain tumors in different populations [10, 11]. The IL-4R gene is also highly polymorphic and single nucleotide polymorphisms of IL-4R have been identified for their implication in cancer on three polymorphic sites, 175V (rs1805010), Q576R (rs1801275) and S503p (rs1805015) [12]. Many studies suggest that these polymorphisms are involved in the etiology of various cancers, including renal cell carcinoma, gastric cancer, pancreatic cancer, brain tumors and cervical cancer [13-17].
Although brain glioma is a major concern in Iraq, no published information is currently available to demonstrate the association between polymorphisms in the IL-4 and IL-4R genes with glioma development and progression. In addition, these mutations may also diminish or abrogate the host tumor surveillance. Studying these SNPs may be beneficial in developing possible diagnostic and interventional strategies for these patients. To this end, in the present study, we investigated the association of SNPs in IL-4 and the IL-4R genes and the systemic level of IL-4 with the incidence of glioma in Iraqi patients.
Materials and Methods
Five hospitals in Iraq, Neurosurgery Hospital, Neuroscience Hospital, Al-kadimiya Teaching Hospital, Al-Fallujah Hospital, and a Private Nursing Home Hospital near Baghdad were included in this study. Patients attending these hospitals to undergo surgical resection of brain tumors from January 2012 to January 2013 were included in this study. Ethical clearance to conduct the research was obtained from these hospitals separately. Selection of patients was accomplished after the assistance of surgeons in the hospitals. One hundred patients were selected for this study. All patients had glioma brain tumors of different grade and stages (45 men and 55 women) and with a mean age of 40.2 years (range between 4-70). Forty controls were selected randomly from apparently healthy individuals (27 men and 13 women) with a mean age of 33.9 years (range between 21- 55). Individuals previously diagnosed with cancer at the time of enrollment, were excluded from this group. Data were collected through direct interview with the participants, and by seeking patient's hospital record as well as previous medical reports. Patient's claims were followed as an alternative source of information when his/her previous medical reports were not available. These data included age, previous and current occupation, smoking, drinking, residence, and first relative family history of brain cancer.
Blood Samples
Five-milliliters (ml) of blood were taken from normal healthy controls and patients before the initiation of chemotherapy or radiation therapy. 2 mls were kept in an EDTA tube (for the SNPs study) at -20oC for further analysis. The remaining 3 mls were collected in an uncoated tube and serum was obtained by centrifugation then stored at -80oC until used for estimation of serum levels of IL-4.
DNA Extraction and Genotyping
DNA was extracted from each blood sample using the gSYNCTM DNA Mini Kit with Whole Blood Protocol (Geneaid/ Korea), according to the manufacturer's protocol. The primers used for IL-4 gene amplification were 5'-GATTTGCAGTGACAATGTGAG-3' as the forward primer and 5'- TCCTATGCTGAAACTTTGTAG-3' as the reverse primers, while those for the IL-4R gene were 5'-CTTACCGCAGCTTCAGCGAC-3'as the forward perimer and 5' CACAGTGGTTGGCTCAGAGA-3' as the reverse primer. Template DNA (10 µl) from each sample and primers (5 µL of each) were added to the PCR master-mix (Bioneer/Korea) (50 µl). The mixture was then put in a shaker and spinner for 10 cycles for mixing. After mixing, the master-mix tubes were transferred to the thermocycler (MyGenie 32 thermal block/Bioneer/Korea). The PCR protocol for IL-4 gene amplification was as follows: initial denaturation at 95°C for 5 min, followed by 30 amplification cycles of 94°C for 30 seconds (denaturation), 60°C for 30 seconds (primer annealing) and 72°C for 1 minute (extension). The final extension was at 72°C for 10 min. The PCR protocol for IL-4R gene amplification was as follows: initial denaturation at 95°C for 5 min, followed by 35 amplification cycles of 94°C for 30 seconds (denaturation), 57°C for 30 seconds (primer annealing) and 72°C for 1 minute (extension). The final extension was at 72°C for 10 min.
PCR Product purification
PCR product purification was carried using the gSYNCTM PCR Product purification Mini Kit (Geneaid/ Korea) according to the manufacturer's instructions.
Measurement of DNA Concentration of Purified PCR product
The DNA concentration of the Purified PCR products was measured using a Nanodrop/UVS-99 (ACTGene/USA). All products had a concentration of more than 100ng/mL and DNA was sequenced.
DNA Sequencing
Polymerase chain reaction products were sent to Bioneer Company/Korea for DNA sequencing. The obtained sequences were aligned using "ClustalW" software with a normal sequence from GenBank (GenBank accession number NC_000005.10 and NC_000016.10 for IL-4 and IL-4R genes respectively) and examined for the presence of polymorphism.
Estimation of serum levels of IL-4
An enzyme-linked immunosorbent assay (ELISA) kit from (Maptech, USA) was used to estimate serum levels of IL-4 in each sample according to the manufacturer's instructions.
Statistical Analysis
The Statistical Package for the Social Sciences (SPSS, version 14) was used for statistical analysis. Risk association between the genotype and brain tumor glioma susceptibility and the distribution of different alleles between patients and controls were estimated by the calculation of adjusted odds ratio and 95% confidence intervals using binary logistic regression. Chi- Square analysis was used for testing the deviation from Hardy-Weinberg equilibrium and serum levels of IL-4. All p-values < 0.05 were considered statistically significant.
Results
Characteristics of the participants in this study
Table 1 shows the characteristics and risk factors in this study of glioma patients and healthy controls. Although there was some disparity in the mean of age between the patient and control groups, a logistic regression test did not reveal any significant association between age and glioma development (OR= 1.408, 95%CI=0.581- 3.415, p=0.448). Similarly, gender does not appear to be a risk factor for glioma as there were no significant differences in the incidence of the disease between male and female subjects in the patient and control groups (OR= 1.408, 95%CI=0.581-3.415).
Risk factors in patients with glioma and controls
| Variables | Cases N=100 | Control N=40 | P-value | OR (95%CI) |
|---|---|---|---|---|
| IL-4 SNP | ||||
| CC | 71 (71%) | 22 (55%) | 0.173 | 1.0 |
| CT | 26 (26%) | 13 (32.5%) | 0.423 | 0.696 (0.287-1.688) |
| TT | 3 (3%) | 5 (12.5%) | 0.053 | 0.19 (0.035-1.02) |
| IL-4R SNP | ||||
| TT | 70(70%) | 17(42.5%) | 0.10 | 1.0 |
| CT | 25 (25%) | 15(37.5%) | 0.042 | 0.405 (0.17-0.969) |
| CC | 5(5%) | 8(20%) | 0.008 | 0.147(0.036-0.60) |
| Mean age in years (SD) | 35.08 (16.2) | 30.1 (12.24) | 0.448 | 1.408 (0.581- 3.415) |
| Family history | 0.062 | |||
| No | 67(67%) | 34(85%) | 1.0 | |
| Yes | 33(33%) | 6 (15%) | 2.615(0.953-7.377) | |
| Sex | 0.4448 | |||
| Female | 43(43%) | 16(40%) | 1.0 | |
| Male | 57(57%) | 24(60%) | 1.408(0.581-3.415) | |
| Smoking | ||||
| Never | 61(61%) | 30(75%) | 0.123 | 1.0 |
| Ex-smoker | 17(17%) | 6 (15%) | 0.318 | 1.77(0.83-4.87) |
| Current | 22(22) | 4(10%) | 1.92(0.612-5.828) |
In contrast, there was a positive association (although not significant) between family history of cancer among one or more individual first relatives and the incidence of glioma (OR= 2.615, 95%CI=0.953-7.377, p=0.062 (Table 1). Smoking status has been categorized into three categories: never, ex-smokers and current smokers. A larger percentage of subjects who never smoked were among the control group (75% compared to 61%), while 15% of control subjects were ex-smokers compared to 17% of glioma patients, and 10% of the control group were current smokers compared to 23% of glioma patients. Nevertheless, there was no significant association between smoking and glioma (OR= 1.77, 95% CI=0.83-4.87 and OR=1.92, 95% CI=0.612-5.828 for ex- smokers and current smokers respectively) (Table 1).
Polymorphisms in the IL-4 and IL-4R genes are associated with glioma patients
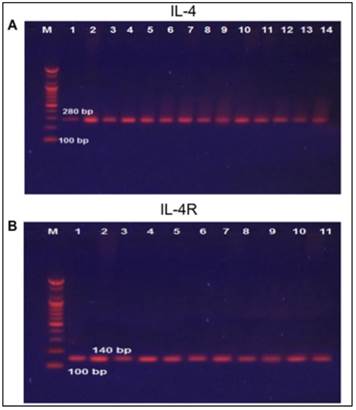
IL-4 acts in conjunction with IL-13 in the regulation of allergic inflammation [18]. It has been shown that single nucleotide polymorphisms in the IL-4 gene are associated with the incidence of glioma [19]. Further, it has been reported that polymorphisms in the IL-4R gene are associated with better survival in glioma patients [19]. To demonstrate any such changes in the present study, we initially demonstrated the generation of PCR products using gel electrophoresis for portions of the IL-4 and IL-4R genes in Fig. 1A and B, respectively.
Gel electrophoresis for (A) IL-4 PCR product visualized under UV light. M, 1000 bp marker, lane 1-10: from blood of glioma patients, lane 11-14: from blood of control group. The size of product is 280 bp and (B) IL-4R PCR product visualized under UV light. M, 1000 bp marker, lane 1-9: from blood of glioma patients, lane 10-11: from blood of control group. The size of product is 140 bp.
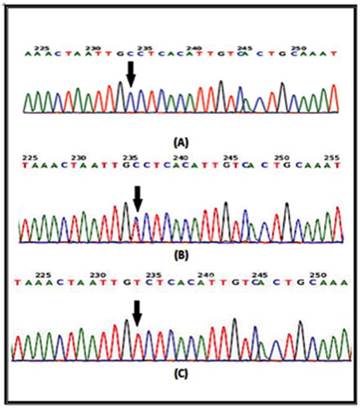
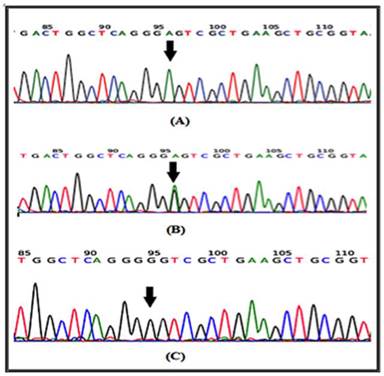
In a recent report, several SNPs in the IL-4 gene have been identified [20]. It has been shown that at least one of the IL-4-589C>T (rs 2243250) allele shows functional polymorphisms. Furthermore, this allele has been shown to increase IL-4 transcriptional activity [21]. Here the sequencing involved portion of the IL-4 gene extending from nucleotide number 132009730 to nucleotide number132009449 on chromosome 5. Similarly, for the IL-4R gene, the sequencing involved part of the gene extending from nucleotide number 27374160 to nucleotide number 27374299 on chromosome 16. The results of the DNA sequencing of the PCR product for the portion of the IL-4 gene (forward strand) clearly demonstrates the IL-4 polymorphism. The arrow indicates that the position of SNP rs2070784 represents (a), the genotype CC; (b) TC and (c) TT (Fig. 2). Similarly, DNA sequencing for part of the IL-4R gene (reverse strand) also shows polymorphisms at position SNP rs1805015 representing (a) the genotype TT; (b) TC and (c) CC (Fig. 3).
SNP rs2070784 within the IL-4 gene is protective in glioma patients
SNP rs2070784 (IL4) has three genotypes: CC, CT, and TT in both glioma and control groups (Table 1). In glioma patients, these genotypes account for 71 (71%), 26 (26%), and 3 (3%) respectively, compared with 22 (55%), 13 (32.5%), and 5 (12.5%), respectively in the control group. Although it appears that the T allele in CT and TT genotypes is associated with a protective effect against glioma, the association is insignificant for the CT genotype (OR=0.696, 95%CI=0.287-1.688, p=0.423), but very close to significance (p=0.053) for the TT genotype (OR=0.19, 95%CI=0.035-1.02).
DNA sequencing from portions of the IL-4 gene from the forward strand are shown. The arrow indicates the position of SNP rs2070784 a: represents the genotype CC; b: TC and c: TT.
DNA sequencing for a portion of the IL-4R gene from the reverse strand is shown. The arrow indicates the position of SNP rs1805015 a: represents the genotype TT; b: TC and c: CC.
SNP rs1805015 (IL-4R) is protective in glioma patients
SNP rs1805015 within the IL-4R gene has three genotypes: TT, CT, and CC in both patient and control groups (Table 1). In glioma patients, these genotypes account for 70 (70%), 25 (25%), and 5 (5%), respectively, when compared with 17 (42.5%), 15 (37.5%), and 8 (20%), respectively in subjects within the control group. Here, significance of the association between the protective effect of allele T and glioma formation is more obvious. For the genotype CT, the OR=0.405, 95%CI =0.17-0.969, p= 0.042, which means that individuals carrying this genotype are 2.47-fold (1/0.405) less likely to acquire gliomas compared with individuals carrying the TT genotype. The association is more prominent with the CC genotype where OR= 0.147, 95% CI=0.036-0.60, p=0.008, which means that individuals carrying CC genotypes are 6.8-fold (1/0.147) less likely to acquire glioma compared with individuals carrying the TT genotype.
Allele analysis confirmed the aforementioned results using Chi-square analysis for examining allele distribution and the results indicate that both SNPs meet the stringent Weinberg equilibrium. For SNP rs2070784, frequencies of the C allele (wild-type) among patient and control groups are 84% and 71.25%, respectively, while frequency of the T allele (mutant) among patients and control subjects were significantly different (p=0.02) at 16% and 28.75%, respectively. Similarly, the frequency of the wild type T allele at rs1805015 among glioma patients and controls were 82.5% and 61.25%, respectively, while the frequency of the mutant C allele was significantly different (p=0.025) with 38.75% among the control subjects versus 17.5% in glioma patients, which indicates the protective effect of this allele in glioma patients (Table 2).
Systemic serum concentration of Interleukin-4
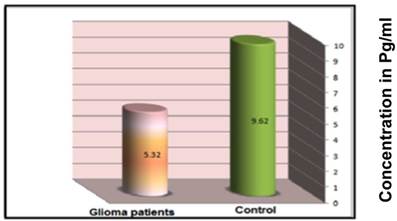
Glioma patients and healthy controls were examined for circulating IL-4 concentration in their sera. The results clearly indicate a higher serum concentration of IL-4 among control group subjects (9.62±0.27pg/mL) than that of glioma patients (5.32±0.39 pg/mL) at a significance of p=0.028 (Figure 4). These data strongly suggest that higher concentrations of IL-4 are protective against glioma.
Allele frequency between patients and controls for the SNPs
| SNPs | Alleles | Cases | Control | χ2 | p-value |
|---|---|---|---|---|---|
| IL-4 | C (wild) | 168(84%) | 57 (71.25%) | 5.585 | 0.02 |
| rs2070784 | T (mutant) | 32 (16%) | 23 (28.75%) | ||
| IL-4R | T (wild) | 165(82.5%) | 49(61.25%) | 14.32 | 0.0025 |
| rs180505 | C (mutant) | 35(17.5%) | 31(38.75%) |
Mean serum concentration of IL-4 (pg/ml) in glioma patients and control group subjects as described in the materials and methods.
Discussion
Studies have reported an association between glioma susceptibility and various inflammatory factors that either modulate the immune response or serve as a surrogate for immune dysfunction (e.g., polymorphisms in immune genes, immunoglobulin E levels (IgE), atopic conditions and use of antihistamine) [22, 23]. Interleukin-4 is one of the key players in an immune response and dysregulation in the production of this cytokine and/or its receptor may strongly influence the immune system with subsequent decreased or increased susceptibility to glioma [24]. The results from our current study clearly indicate a protective role of SNPs in the IL-4 (rs2070784) and IL-4R (rs1805015) gene against glioma. These results are in agreement with a previously published study [19].
While the precise mechanism of this protection is not fully understood, it is thought that SNP rs2070784 (C-33T) results an increase in IL-4 production [25]. In the present study, we also noted a significant increase in serum levels of IL-4 among control versus glioma patients, while the SNP rs1805015 (S503P) is thought to amplify the action of IL-4. Therefore, it is reasonable to believe that these SNPs increase production and amplify IL-4 levels, thereby influencing glioma susceptibility in the patients. For the SNP C-33T in the IL-4 gene, the exact mechanism for this increment (change in IL-4 production) is not known. However, since the SNP is located within 5'UTR of the gene, it may be possible that gene alterations may influence gene transcription and/or mRNA stabilization, perhaps involving specific functional motifs such as conformational loops and hairpins that play an important role in function and transcription control, especially in 5'UTR [26]. Another study suggests that the locus containing C-33T is part of a cAMP responsive element binding protein (CREB)-binding site and CREB is a regulatory protein involved in the regulation of IL-4 expression through the modification of the CREB-binding site [27]. Hence, the SNP may result in increased IL-4 production through an increase in the response to cAMP.
The exact mechanism by which SNP S503P in the IL-4R gene amplifies the action of IL-4 is not well known. IL-4 binds with the type I IL-4Rα complex composed of the IL-4Rα chain, common (γC) chain, and/or the type II IL-4R complex composed of the IL-4rα chain and IL-13Rα1 chain [28]. Signal transduction downstream of both of these receptors occurs via signal transducer and activator of transcription-6 (STAT6) and the insulin receptor substrate pathways [17]. STAT6 promotes transcription of GATA3 (a TH2-cell inducer) and MHC II to induce IgE class switching in B cells [29]. Furthermore, the amino acid encoded by SNP rs1805015 is located in the intracellular domain of the IL-4R close to the substrate-binding site, and it is part of the insulin-IL-4 receptor (IRS) motif. This motif contains a tyrosine residue at position 495, which is phosphorylated by Janus Kinase I (JAK1) [30]. This amino is crucial for binding and tyrosine phosphorylation of IRS-1 and IRS-2, which consequently initiate signal transduction cascade towards cell proliferation [31]. Several studies have also suggested that stimulation in IL-4R and STAT6 signaling results in enhanced STAT6 activity, which may cause an increase in TH2 cell differentiation [32], which is the case in the presence of SNP rs1805015 in the IL-4R gene.
Increased plasma concentrations of IL-4 through several mechanisms can influence the immune status of an affected individual and result in many phenotypes beyond the scope of the immune system. Collectively these effects may alter susceptibility to brain tumor development. It has been shown that cytokines suppress DNA synthesis and cell proliferation in normal astrocytes and low-grade astrocytoma cell lines, but not in GMB cell lines [33]. These results suggest that IL-4 may interfere with the progression from low to high-grade glioblastoma [34]. In contrast, growth-inhibitory properties of mouse IL-4 on implanted glioma cell lines limits its ability to promote eosinophil infiltration and to inhibit angiogenesis [35]. Supporting the latter assumption is the observation that IL-4 blocks corneal neo-vascularization by fibroblast growth factor in mice and inhibits the migration of human microvascular cells [36].
The most plausible theory for the protective role of high concentration of IL-4 against glioma is its role in allergy, which is well documented and inversely associated with glioma risk [37]. IL-4 is implicated in many cellular and biochemical events that account for allergy and it regulates Th2 cells, mast cells, eosinophils, neutrophils, goblet cell hyperplasia, IgE production, adhesion molecules, chemokines, and airway hyperresponsiveness. It has been shown that administration of IL-4 (or IL-13) to mice induces asthma symptoms [18]. Towards this, the SNP C-33T was particularly found to be associated with increased risk of asthma [38]. If such an association is present, it is reasonable to assume that eliminating the allergy from those who suffer from this disease may increase the incidence of glioma among them. In a recent study it has been shown that oral administration of antihistamine is significantly associated with glioma risk among individuals reporting a history of allergies/asthma [39]. However, it is not clear whether allergies can reduce glioma risk or their relative absence in glioma patients is merely a reflection of the immunosuppressive effect of the tumor [40]. Additional supporting evidence for the inverse association between allergy and glioma is that serum levels of IgE in a clinical trial on the IgE blocking drug omalizumab showed a higher rate of solid tumors in the treated group compared to the control group [41]. Furthermore, glioma cases exhibit lower IgE levels than population-based frequency-matched controls [7], and patients with higher IgE levels have a better antitumor defense or less aggressive tumors with weaker anti-immunologic effects [42].
These observations and results from the present study suggest that SNPs C-33T and S503P may have a protective role against glioma through induction of an allergic status. However, more detailed studies in relation to allergic status are required for more confidence in this conclusion.
Acknowledgements
We thank Dr. Haider and Dr. Ahmad Adnan in the Neuroscience Hospital for help in collection of patient samples and Dr. Qasim Sharhan for critical comments and technical assistance. We are grateful to Dr. Haider Sabah for critical review of this manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kyritsis AP, Bondy ML, Rao JS, Sioka C. Inherited predisposition to glioma. Neuro-Oncology. 2010;12(1):104-113
2. Prasad G, Haas-Kogan DA. Radiation-induced gliomas. Expert Review of Neurotherapeutics. 2009;9(10):1511-1517
3. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathologica. 2005;109(1):93-108
4. Davis F, Il'yasova D, Rankin K, McCarthy B, Bigner DD. Medical diagnostic radiation exposures and risk of gliomas. Radiation Research. 2011;175(6):790-796
5. Melin B. Genetic causes of glioma: new leads in the labyrinth. Curr Opin Oncol. 2011;23(6):643-647
6. Lohr J, Ratliff T, Huppertz A, Ge Y, Dictus C, Ahmadi R, Grau S, Hiraoka N, Eckstein V, Ecker RC. et al. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-beta. Clin Cancer Res. 2011;17(13):4296-4308
7. Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, Gohlke H, Wagenpfeil S, Ollert M, Ring J. et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4(8):e1000166
8. Mearns H, Horsnell WG, Hoving JC, Dewals B, Cutler AJ, Kirstein F, Myburgh E, Arendse B, Brombacher F. Interleukin-4-promoted T helper 2 responses enhance Nippostrongylus brasiliensis-induced pulmonary pathology. Infection & Immunity. 2008;76(12):5535-5542
9. Babula O, Lazdane G, Kroica J, Linhares IM, Ledger WJ, Witkin SS. Frequency of interleukin-4 (IL-4) -589 gene polymorphism and vaginal concentrations of IL-4, nitric oxide, and mannose-binding lectin in women with recurrent vulvovaginal candidiasis. Clin Infect Dis. 2005;40(9):1258-1262
10. Brenner AV, Butler MA, Wang SS, Ruder AM, Rothman N, Schulte PA, Chanock SJ, Fine HA, Linet MS, Inskip PD. Single-nucleotide polymorphisms in selected cytokine genes and risk of adult glioma. Carcinogenesis. 2007;28(12):2543-2547
11. Jin T, Li X, Zhang J, Wang H, Geng T, Li G, Gao G, Chen C. Genetic association between selected cytokine genes and glioblastoma in the Han Chinese population. BMC Cancer. 2013;13:236
12. Wang XW, Boisselier B, Rossetto M, Marie Y, Idbaih A, Mokhtari K, Gousias K, Hoang-Xuan K, Delattre JY, Simon M. et al. Prognostic impact of the isocitrate dehydrogenase 1 single-nucleotide polymorphism rs11554137 in malignant gliomas. Cancer. 2013;119(4):806-813
13. Olson SH, Orlow I, Simon J, Tommasi D, Roy P, Bayuga S, Ludwig E, Zauber AG, Kurtz RC. Allergies, variants in IL-4 and IL-4R alpha genes, and risk of pancreatic cancer. Cancer Detect Prev. 2007;31(5):345-351
14. Castro FA, Haimila K, Sareneva I, Schmitt M, Lorenzo J, Kunkel N, Kumar R, Forsti A, Kjellberg L, Hallmans G. et al. Association of HLA-DRB1, interleukin-6 and cyclin D1 polymorphisms with cervical cancer in the Swedish population--a candidate gene approach. Int J Cancer. 2009;125(8):1851-1858
15. Mohan S, Mohanasenthil, Paul SF, Shroff S, Venkatesan V. Interleukin-4-receptor alpha gene polymorphism and the risk of renal cell carcinoma in a South Indian population. Asian Pac J Cancer Prev. 2009;10(2):295-298
16. Chu H, Wang M, Yan F, Zhong D, Shi D, Ma L, Pan X, Qin C, Yin C, Zhang Z. Polymorphisms in the IL-13 and IL-4R genes are associated with the development of renal cell carcinoma. Ann Oncol. 2012;23(8):2114-2121
17. Ford AQ, Heller NM, Stephenson L, Boothby MR, Keegan AD. An atopy-associated polymorphism in the ectodomain of the IL-4R(alpha) chain (V50) regulates the persistence of STAT6 phosphorylation. J Immunol. 2009;183(3):1607-1616
18. Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, Sheppard D, Mohrs M, Donaldson DD, Locksley RM. et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282(5397):2261-2263
19. Scheurer ME, Amirian E, Cao Y, Gilbert MR, Aldape KD, Kornguth DG, El-Zein R, Bondy ML. Polymorphisms in the interleukin-4 receptor gene are associated with better survival in patients with glioblastoma. Clinical Cancer Research. 2008;14(20):6640-6646
20. Kabesch M, Tzotcheva I, Carr D, Hofler C, Weiland SK, Fritzsch C, von Mutius E, Martinez FD. A complete screening of the IL-4 gene: novel polymorphisms and their association with asthma and IgE in childhood. J Allergy Clin Immunol. 2003;112(5):893-898
21. Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish L. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clinical & Experimental Allergy. 1995;25(Suppl 2):74-78 discussion 95-76
22. Bush A, Pedersen S, Hedlin G, Baraldi E, Barbato A, de Benedictis F, Lodrup Carlsen KC, de Jongste J, Piacentini G. Pharmacological treatment of severe, therapy-resistant asthma in children: what can we learn from where? Eur Respir J. 2011;38(4):947-958
23. Busse WW. Asthma diagnosis and treatment: filling in the information gaps. J Allergy Clin Immunol. 2011;128(4):740-750
24. Israel E, Chinchilli VM, Ford JG, Boushey HA, Cherniack R, Craig TJ, Deykin A, Fagan JK, Fahy JV, Fish J. et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364(9444):1505-1512
25. Almomani B, Hawwa AF, Millership JS, Heaney L, Douglas I, McElnay JC, Shields MD. Can certain genotypes predispose to poor asthma control in children? A pharmacogenetic study of 9 candidate genes in children with difficult asthma. PLoS One. 2013;8(4):e60592
26. Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122(Pt 12):1933-1937
27. Gervaziev YV, Kaznacheev VA, Gervazieva VB. Allelic polymorphisms in the interleukin-4 promoter regions and their association with bronchial asthma among the Russian population. Int Arch Allergy Immunol. 2006;141(3):257-264
28. Callard RE, Matthews DJ, Hibbert L. IL-4 and IL-13 receptors: are they one and the same? Immunol Today. 1996;17(3):108-110
29. Corry DB, Folkesson HG, Warnock ML, Erle DJ, Matthay MA, Wiener-Kronish JP, Locksley RM. Interleukin 4, but not interleukin 5 or eosinophils, is required in a murine model of acute airway hyperreactivity. J Exp Med. 1996;183(1):109-117
30. Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, Deichmann KA. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology. 1999;96(3):365-371
31. Keegan AD, Nelms K, White M, Wang LM, Pierce JH, Paul WE. An IL-4 receptor region containing an insulin receptor motif is important for IL-4-mediated IRS-1 phosphorylation and cell growth. Cell. 1994;76(5):811-820
32. Stephenson L, Johns MH, Woodward E, Mora AL, Boothby M. An IL-4R alpha allelic variant, I50, acts as a gain-of-function variant relative to V50 for Stat6, but not Th2 differentiation. Journal of Immunology. 2004;173(7):4523-4528
33. Barna BP, Estes ML, Pettay J, Iwasaki K, Zhou P, Barnett GH. Human astrocyte growth regulation: interleukin-4 sensitivity and receptor expression. J Neuroimmunol. 1995;60(1-2):75-81
34. Louis DN. A molecular genetic model of astrocytoma histopathology. Brain Pathol. 1997;7(2):755-764
35. Saleh M, Wiegmans A, Malone Q, Stylli SS, Kaye AH. Effect of in situ retroviral interleukin-4 transfer on established intracranial tumors. Journal of the National Cancer Institute. 1999;91(5):438-445
36. Volpert OV, Fong T, Koch AE, Peterson JD, Waltenbaugh C, Tepper RI, Bouck NP. Inhibition of angiogenesis by interleukin 4. J Exp Med. 1998;188(6):1039-1046
37. Linos E, Raine T, Alonso A, Michaud D. Atopy and risk of brain tumors: a meta-analysis. J Natl Cancer Inst. 2007;99(20):1544-1550
38. Yang HJ. Association between the interleukin-4 gene C-589T and C+33T polymorphisms and asthma risk: a meta-analysis. Arch Med Res. 2013;44(2):127-135
39. Amirian ES, Marquez-Do D, Bondy ML, Scheurer ME. Antihistamine use and immunoglobulin E levels in glioma risk and prognosis. Cancer Epidemiol. 2013;37(6):908-912
40. Schwartzbaum JA, Huang K, Lawler S, Ding B, Yu J, Chiocca EA. Allergy and inflammatory transcriptome is predominantly negatively correlated with CD133 expression in glioblastoma. Neuro Oncol. 2010;12(4):320-327
41. Schwartzbaum J, Ahlbom A, Malmer B, Lonn S, Brookes AJ, Doss H, Debinski W, Henriksson R, Feychting M. Polymorphisms associated with asthma are inversely related to glioblastoma multiforme. Cancer Res. 2005;65(14):6459-6465
42. Wrensch M, Wiencke JK, Wiemels J, Miike R, Patoka J, Moghadassi M, McMillan A, Kelsey KT, Aldape K, Lamborn KR. et al. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006;66(8):4531-4541
Author contact
![]() Corresponding author: Udai P. Singh, Ph.D., Department of Pathology, Microbiology and Immunology, University of South Carolina, Columbia, SC 29208. Phone: (803) 216-3423; Fax: (803) 216-3413; Email: udai.singhsc.edu
Corresponding author: Udai P. Singh, Ph.D., Department of Pathology, Microbiology and Immunology, University of South Carolina, Columbia, SC 29208. Phone: (803) 216-3423; Fax: (803) 216-3413; Email: udai.singhsc.edu

 Global reach, higher impact
Global reach, higher impact