Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(11):1129-1132. doi:10.7150/ijms.9487 This issue Cite
Short Research Communication
Relationship between European Mitochondrial Haplogroups and Chronic Renal Allograft Rejection in Patients with Kidney Transplant
1. Unidad de Infección Viral e Inmunidad, Centro Nacional de Microbiología. Instituto de Salud Carlos III, Majadahonda, Spain.
2. Departamento de Anestesiología y Reanimación, Hospital Clínico Universitario, Valladolid, Spain.
3. Unidad de Investigación Biomédica. Hospital Clínico Universitario-IECSCYL, Valladolid, Spain.
Received 2014-4-24; Accepted 2014-7-15; Published 2014-8-17
Abstract
Mitochondrial DNA variants may contribute to differences in mitochondrial function, leading to an altered immune system. The aim of this study was to analyze the relationship between mtDNA haplogroups and the development of chronic allograft dysfunction in patients with kidney transplant. A retrospective observational study was carried out on 261 patients who received kidney transplant (114 had stable transplant and 147 patients developed chronic allograft dysfunction). DNA samples were genotyped for 14 mtDNA polymorphisms by using Sequenom's MassARRAY platform (San Diego, CA, USA). Only European white patients within the N macro-cluster were included. Patients with haplogroups V (odds ratio (OR)=0.32; p=0.037) and J (OR=0.36; p=0.038) showed lower odds for developing CRAD than patients with haplogroup H. After adjusting for the most significant variables, haplogroups V and J tended to statistical significance (p=0.091 and p=0.067 respectively). This is a preliminary study in which mtDNA haplogroups seem to be implicated in susceptibility or protection for developing chronic allograft dysfunction.
Keywords: kidney transplant, allograft rejection, SNPs, mitochondrial haplogroups, mtDNA
INTRODUCTION
Kidney transplant (KT) is the best therapeutic alternative for most causes of chronic renal failure [1, 2], where avoiding the chronic renal allograft dysfunction (CRAD) is essential [3]. The CRAD is a multifactorial pathological entity characterized by a progressive decrease in glomerular filtration rate, generally associated with proteinuria and arterial hypertension [4]. Nowadays, KT outcomes are not completely understood, but there is growing evidence that mitochondrial genetic background may contribute to differences between individuals in this complex disease.
Mitochondria are the main energy-producing organelles in mammalian cells, but they also play a central role in cell injury and death signaling. In addition, mitochondria dysfunction plays a critical role in the pathogenesis of kidney diseases and it has been related to renal failure [5]. Mitochondrial DNA (mtDNA) is highly polymorphic due to the fact that specific mtDNA polymorphisms have been acquired throughout human history. Based on these polymorphisms, we are able to classify the human population into a number of discrete mitochondrial clades or haplogroups[6]. Variations in mtDNA have been directly associated with susceptibility to chronic diseases [7], and contribute to differences in mitochondrial function, such as reactive oxygen species (ROS) production and oxidative phosphorylation capacity [8, 9], affecting the function of the immune system [10]. In this setting, mtDNA haplogroup H has been previously related to transplant-related complications [11]. However, the influence of mtDNA haplogroups on the development of allograft failure has not been studied so far. Thus, the aim of this study was to analyze the association of mtDNA haplogroups with the development of CRAD in patients with KT.
PATIENTS AND METHODS
A retrospective observational study was carried out on 261 patients who received KT in the “Hospital Clínico Universitario” in Valladolid (Spain), between 1995 and 2008. Patients were classified into two groups according to their kidney graft outcomes: a) Non-CRAD: patients with stable transplant or absence of CRAD; b) CRAD: patients who developed CRAD. CRAD was considered as any decrease ≥30% of the inverse of creatinine (1/Cr) after the third month of transplantation, using as baseline the highest value of 1/Cr in the first three months after transplantation, as previously described by Kasiske et al [12]. Demographic and clinical data were obtained from medical records. Patients who were younger than 18 years and/or had unavailable DNA sample were ruled out. Written consent was obtained from all patients and the Ethics Committee approved the study.
DNA samples were genotyped for 14 mtDNA polymorphisms that define European mtDNA haplogroups (adapted from Hendrickson et al [13]) at the Spanish National Genotyping Center (CeGen; http://www.cegen.org/) by using Sequenom's MassARRAY platform (San Diego, CA, USA) with the iPLEX® Gold assay design system. Our study included only European white participants within the N macro-cluster, which is ancestral to almost all the European and many of the Eurasian haplogroups [6]. These subjects (N macro-cluster) were further parsed into the most common European major-haplogroups or clusters (HV, U, JT and IWX,) and haplogroups (H, V, pre-V, J, T, I, W and X).
Chi-squared/Fisher tests and Mann-Whitney U tests were used for comparison of variables across groups. Logistic regression was used to assess the relationship between mtDNA haplogroups and development of CRAD. For multivariate analysis, age and gender of recipients and donors, HLA mismatches and cold ischemia time were used to select the most significant covariates by a stepwise algorithm (at each step, factors are considered for removal or entry: a p-value for entry of 0.20 and exit of 0.15). Statistical analysis was performed by SPSS 19.0 software (SPSS INC, Chicago, IL, USA). All tests were two-tailed with p-values <0.05 considered significant.
RESULTS
Table 1 shows the clinical characteristics of 261 patients included in our study: 114 were within Non-CRAD group, and 147 within CRAD group.
Clinical characteristics of patients with renal allograft transplant.
| Characteristics | Non-CRAD | CRAD | p-value |
|---|---|---|---|
| No. | 114 | 147 | |
| Receptor | |||
| Age a | 50 (42.7-57.2) | 52 (42-60) | 0.250 |
| Male b | 51 (44.7) | 49 (33.3) | 0.060 |
| Primary disease b | |||
| Glomerulonephritis | 32 (28.1) | 43 (29.3) | 0.780 |
| Arterial hypertension | 14 (12.3) | 14 (9.5) | 0.499 |
| Diabetic nephropathy | 8 (7) | 11 (7.5) | 0.862 |
| Tubulointerstitial nephritis | 9 (7.9) | 18 (12.2) | 0.237 |
| Obstructive uropathy | 6 (5.3) | 5 (3.4) | 0.543 |
| Vascular causes | 4 (3.5) | 0 (0) | 0.036 |
| Polycystic kidney disease | 18 (15.8) | 17 (11.6) | 0.342 |
| Others/Unknown | 23 (20.2) | 39 (26.5) | |
| Donor | |||
| Age a | 46 (28.7-55) | 53 (39-60) | 0.002 |
| Male b | 78 (68.4) | 98 (66.7) | 0.764 |
| Cause of death b | |||
| CVA | 55 (48.2) | 85 (57.8) | 0.124 |
| TBI | 11 (9.6) | 13 (8.8) | 0.823 |
| Others | 48 (42.1) | 49 (33.3) | |
| Transplant factors | |||
| HLA A mismatches b | 0.510 | ||
| 0 | 65 (57) | 74 (50.7) | |
| 1 | 45 (39.5) | 68 (46.6) | |
| 2 | 4 (3.5) | 4 (2.7) | |
| HLA B mismatches b | 0.969 | ||
| 0 | 77 (67.5) | 99 (67.8) | |
| 1 | 35 (30.7) | 45 (30.8) | |
| 2 | 2 (1.8) | 2 (1.4) | |
| HLA DR mismatches b | 0.834 | ||
| 0 | 13 (11.4) | 16 (11) | |
| 1 | 91 (79.8) | 120 (82.2) | |
| 2 | 10 (8.8) | 10 (6.8) | |
| Cold ischemia time (minutes) a | 693.5 (600.0-913.7) | 849.0 (600-1023) | 0.023 |
CRAD: chronic renal allograft dysfunction; CVA: cerebrovascular accident; TBI: traumatic brain injury.
a Median (interquartile rank). b Absolute number (percentage). NA: not applicable
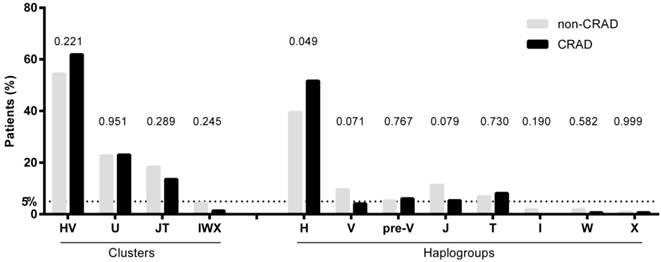
Figure 1 shows the frequencies of mtDNA haplogroups according to CRAD. Patients with CRAD showed higher frequency of haplogroup H (p= 0.049) and a lower tendency of haplogroups V and J (p=0.071 and p=0.079, respectively). The cluster IWX and haplogroups I, X, and W had frequencies of less than 5%, and were discarded for the association analysis.
Table 2 shows the likelihood of developing CRAD according to mtDNA haplogroups. Patients with haplogroups V (odds ratio (OR)= 0.32; p= 0.037) and J (OR= 0.36; p= 0.038) had lower odds of developing CRAD than patients with haplogroup H. After adjusting for the most significant variables, haplogroups V and J tended to statistical significance (p=0.091 and p=0.067 respectively).
Likelihood of developing chronic renal allograft rejection (CRAD) according to European haplogroups. (a) The most significant covariates were selected by a stepwise algorithm. Significant values are marked in bold.
| Unadjusted | Adjusted (a) | |||
|---|---|---|---|---|
| OR (95%CI) | p-value | OR (95%CI) | p-value | |
| Clusters | ||||
| HV | Reference | - | Reference | - |
| U | 0.89 (0.49; 1.63) | 0.708 | 0.95 (0.50; 1.78) | 0.864 |
| JT | 0.65 (0.32; 1.30) | 0.221 | 0.65 (0.32; 1.32) | 0.233 |
| Haplogroups | ||||
| H | Reference | Reference | - | |
| V | 0.32 (0.11; 0.93) | 0.037 | 0.38 (0.12; 1.17) | 0.091 |
| Pre-V | 0.89 (0.30; 2.66) | 0.832 | 0.95 (0.30; 2.96) | 0.926 |
| J | 0.36 (0.14; 0.95) | 0.038 | 0.39 (0.14; 1.07) | 0.067 |
| T | 0.89 (0.34; 2.34) | 0.810 | 0.81 (0.29; 2.28) | 0.690 |
Abbreviations: OR, odds ratio; 95%CI, 95% of confidence interval; p-value, statistical significance.
mtDNA haplogroups frequencies of patients with renal allograft according to chronic renal allograft dysfunction (CRAD) outcome. P-values were calculated by using chi-squared or Fisher tests.
DISCUSSION
To our knowledge, this is the first study that investigates the association between the European mtDNA and the development of CRAD in KT patients. In our study, haplogroup H showed a tendency to worsen the outcome of renal allograft, while haplogroups V and J showed a trend towards lower odds of CRAD, although statistical significance did not remain after multivariate analysis.
Regarding KT, haplogroup H has been related to complications in renal transplanted patients such as new-onset diabetes in a recent article [11], which could suggest that haplogroup H is a risk factor for a worse outcome. This risk could be explained by the differences in mitochondrial function among different mtDNA haplogroups. In this setting, mitochondrial haplogroup H has been demonstrated to have higher activity in the electron transport chain, producing higher quantities of ATP and ROS than other haplogroups, such as J, which exhibits lower energy efficiency [8, 9]. Besides, an increase of stress oxidative markers, as well as a decrease in antioxidant defense mechanisms in transplant recipients with chronic allograft failure, has been also described [14]. In this regard, the presence of haplogroup H (related to higher ROS production) could contribute to increased oxidative damage, leading to higher probability of renal failure; whereas haplogroup J (lower ROS production) could be related to decreased odds of developing CRAD.
Finally, it should be taken into account that our study is retrospective and has a limited sample size, which might be responsible for the lack of statistical association in the multivariate model. Therefore, this study could be considered as a preliminary study. The mtDNA haplogroups seem to be implicated in susceptibility or protection for developing CRAD. However, further studies with larger sample size and different populations would be interesting in order to corroborate the possible influence of mtDNA on outcome of renal allograft.
Acknowledgements
The authors thank the Spanish National Genotyping Center (CeGen) for providing the SNP genotyping services (http://www.cegen.org). We also acknowledge the patients in this study for their participation.
The authors would also like to thank María Jesús Pérez (Hospital Clinico Universitario de Valladolid) who kindly collected the samples.
Funding/Support
This work was supported by two grants from Instituto de Salud Carlos III (Ref: PI08/0738 and PI11/00245) and from Junta de Castilla y León (Ref: GRS 234/A/08). MAJS, MGF, MGA y DPT are supported by grants from Instituto de Salud Carlos III (CD13/00013, RD12/0017/0024, CD12/00442 and CM12/00043 respectively).
CONFLICT OF INTEREST
The authors do not have a commercial or other association that might pose a conflict of interest.
References
1. Howard K, Salkeld G, White S, McDonald S, Chadban S, Craig JC. et al. The cost-effectiveness of increasing kidney transplantation and home-based dialysis. Nephrology (Carlton). 2009;14:123-32 doi:10.1111/j.1440-1797.2008.01073.x
2. Levey AS, Atkins R, Coresh J, Cohen EP, Collins AJ, Eckardt KU. et al. Chronic kidney disease as a global public health problem: approaches and initiatives - a position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007;72:247-59 doi:10.1038/sj.ki.5002343
3. Campistol JM, Boletis IN, Dantal J, de Fijter JW, Hertig A, Neumayer HH. et al. Chronic allograft nephropathy--a clinical syndrome: early detection and the potential role of proliferation signal inhibitors. Clin Transplant. 2009;23:769-77 doi:10.1111/j.1399-0012.2009.01057.x
4. Pascual J, Perez-Saez MJ, Mir M, Crespo M. Chronic renal allograft injury: early detection, accurate diagnosis and management. Transplant Rev (Orlando). 2012;26:280-90 doi:10.1016/j.trre.2012.07.002
5. Zhan M, Brooks C, Liu F, Sun L, Dong Z. Mitochondrial dynamics: regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int. 2013;83:568-81 doi:10.1038/ki.2012.441
6. Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R. et al. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835-50
7. Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359-407 doi:10.1146/annurev.genet.39.110304.095751
8. Arning L, Haghikia A, Taherzadeh-Fard E, Saft C, Andrich J, Pula B. et al. Mitochondrial haplogroup H correlates with ATP levels and age at onset in Huntington disease. J Mol Med (Berl). 2010;88:431-6 doi:10.1007/s00109-010-0589-2
9. Martinez-Redondo D, Marcuello A, Casajus JA, Ara I, Dahmani Y, Montoya J. et al. Human mitochondrial haplogroup H: the highest VO2max consumer--is it a paradox? Mitochondrion. 2010;10:102-7 doi:10.1016/j.mito.2009.11.005
10. Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D. et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: Implications for population susceptibility to diseases. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2014;1842:208-19 doi:10.1016/j.bbadis.2013.10.016
11. Tavira B, Gomez J, Diaz-Corte C, Llobet L, Ruiz-Pesini E, Ortega F. et al. Mitochondrial DNA haplogroups and risk of new-onset diabetes among tacrolimus-treated renal transplanted patients. Gene. 2014;538:195-8 doi:10.1016/j.gene.2014.01.036
12. Kasiske BL, Andany MA, Danielson B. A thirty percent chronic decline in inverse serum creatinine is an excellent predictor of late renal allograft failure. American journal of kidney diseases: the official journal of the National Kidney Foundation. 2002;39:762-8 doi:10.1053/ajkd.2002.31996
13. Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, Palella FJ. et al. Mitochondrial DNA haplogroups influence lipoatrophy after highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;51:111-6 doi:10.1097/QAI.0b013e3181a324d6
14. Cristol JP, Vela C, Maggi MF, Descomps B, Mourad G. Oxidative stress and lipid abnormalities in renal transplant recipients with or without chronic rejection. Transplantation. 1998;65:1322-8
Author contact
![]() Corresponding author: María Angeles Jiménez Sousa; Centro Nacional de Microbiología, Instituto de Salud Carlos III (Campus Majadahonda); Carretera Majadahonda- Pozuelo, Km 2.2; 28220 Majadahonda (Madrid); Telf.: +34 918 223 278; e-mail: majimenezsousaes
Corresponding author: María Angeles Jiménez Sousa; Centro Nacional de Microbiología, Instituto de Salud Carlos III (Campus Majadahonda); Carretera Majadahonda- Pozuelo, Km 2.2; 28220 Majadahonda (Madrid); Telf.: +34 918 223 278; e-mail: majimenezsousaes

 Global reach, higher impact
Global reach, higher impact