Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(8):810-818. doi:10.7150/ijms.8647 This issue Cite
Review
Expression, Regulation and Function of MicroRNAs in Multiple Sclerosis
1. Institute for Tumor Immunology, Ludong University College of Life Sciences, 186 Hongqi Middle Road, Yantai, Shandong 264025, China
2. Department of Pathology, Microbiology and Immunology, School of Medicine, University of South Carolina, 6439 Garners Ferry Road, Columbia, SC 29209, USA
Received 2014-1-22; Accepted 2014-5-14; Published 2014-6-2
Abstract
MicroRNAs (miRNAs) are single-stranded 19-25 nucleotide-long RNAs and have an important role in post-transcriptional gene silencing. It has been demonstrated that miRNAs are dysregulated in patients with multiple sclerosis (MS). For instance, miR-21, miR-142-3p, miR-146a, miR-146b, miR-155 and miR-326 were up-regulated in both peripheral blood mononuclear cells (PBMCs) and brain white matter lesions from MS patients and mouse model as well. These up-regulated miRNAs may be used as a signature for MS and play critical roles in MS pathogenesis. Moreover, miR-15a, miR-19a, miR-22, miR-210 and miR-223 were up-regulated in both regulatory T cells (Tregs) and other samples such as plasma, blood cells, PBMCs and brain white matter tissues from MS patients, suggesting that these up-regulated miRNAs and Tregs may also play a role in MS pathogenesis. Contrarily, other miRNAs such as miR-15a, miR-15b, miR-181c and miR-328 were down-regulated in MS. Drugs such as interferon-β and glatiramer acetate for MS treatment may regulate miRNA expression and thus have benefits for MS patients. The dysregulated miRNAs such as miR-155 and miR-326 may be used as diagnostic markers and therapeutic targets for MS.
Keywords: miRNA, multiple sclerosis, biomarker, therapeutic target, gene expression, gene regulation
Introduction
Multiple sclerosis (MS) is an autoimmune disease that severely affects people worldwide. MS disease onset usually occurs in young adults, and it is more common in women than men [1, 2]. It hasbeen estimated that a lifetime risk for MS is one out of 400 [1]. MS was discovered about a hundred and fifty years ago; however, neither the etiology nor the mechanism of the disease is fully understood. Moreover, there is also no cure for the disease. The current available treatments have only modest effects and therefore, there is greater need not only for the treatment of MS but also for the understanding the mechanisms of development of the disease.
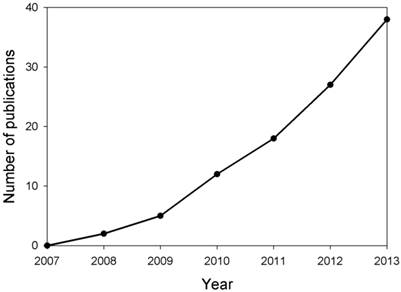
MicroRNAs (miRNAs) are single-stranded 19-25 nucleotide-long RNAs and have an important role in post-transcriptional gene silencing by base pairing with target mRNAs. Recent research has demonstrated that miRNAs are involved in several crucial biological functions, as well as, in a broad spectrum of human diseases such as neurological disorders [3]. It has been reported that miR-29b was up-regulated in memory CD4+ T cells from the peripheral blood of MS patients, however, miR-29b expression was inhibited significantly upon T cell activation in MS patients [4], suggesting that miRNA expression was dysregulated in MS patients and may contribute to MS pathogenesis. Thus, research on the expression, regulation and function of miRNAs in MS has attracted much attention in recent years (Fig. 1). In this paper, we reviewed the recent progress in understanding the roles of miRNAs in MS pathogenesis, and specific miRNAs as biomarkers and therapeutic targets for MS.
Estimation of number of publications about studies on miRNAs in MS yearly from 2007 to 2013. Based on the retrieved papers from the PUBMED database after searching related references by means of miRNA, multiple and sclerosis key words, the number of publications about the studies on miRNAs in MS were calculated.
Expression of miRNAs in MS
Abnormal expression of miRNAs in Peripheral Blood and Plasma
In recent years, there has been a significant focus on understanding the pathogenesis of MS and the role of miRNAs in the pathogenesis of the disease. Both next-generation sequencing and microRNA array analysis demonstrated that miR-16-2-3p was over-expressed in the blood of patients with MS, whereas miR-7-1-3p and miR-20a-5p were down-regulated as compared with healthy controls [5]. An earlier study using miRNA microarray analysis showed that miR-768-3p was up-regulated, whereas 26 miRNAs including miR-15a, miR-15b, miR-16, miR-17, miR-20a, miR-20b, miR-27a, miR-27b, miR-93, miR-98, miR-106a, miR-126, miR-126*, miR-140-5p, miR-211, miR-374a, miR-454, miR-510, miR-579, miR-623, miR-624*, let-7d, let-7f, let-7g and let-7i were significantly down-regulated in peripheral blood of patients with all MS subtypes including primary progressive, secondary progressive and relapsing-remitting disease [6].
miRNA PCR array analysis discovered that plasma levels of 29 miRNAs including miR-9*, miR-20b*, miR-22, miR-29b-2*, miR-30e, miR-30e*, miR-96, miR-134, miR-140-3p, miR-150, miR-151-5p, miR-181b, miR-196a, miR-210, miR-337-5p, miR-342-3p, miR-378, miR-431, miR-449b, miR-451, miR-454, miR-500, miR-502-5p, miR-542-5p, miR-574-3p, miR-671-5p, let-7c, let-7d and let-7f increased in the patients with relapsing-remitting (RR) MS [7]. miRNA array analysis revealed that 6 miRNAs including miR-22, miR-422a, miR-572, miR-614, miR-648 and miR-1826 increased significantly in the plasma of MS patients, but miR-1979 decreased significantly [8]. It has also been reported that miR-155 expression was up-regulated in the serum samples from patients with MS as shown by real-time RT-PCR analysis using the TaqMan miRNA assay [9]. Another study, however, showed that miR-15b, miR-23a and miR-223 were down-regulated in the serum samples from patients with MS as compared with normal controls [10].
Dysregulation of miRNAs in Blood Cells and Peripheral Blood Mononuclear Cells (PBMCs)
miRNA array screening found that 165 miRNAs were significantly deregulated in the blood cells of patients with RRMS, 74 miRNAs (44.8%) were up-regulated, and 91 miRNAs (55.2%) were down-regulated. Among the 10 most significantly deregulated miRNAs, 9 miRNAs including miR-142-3p, miR-145, miR-186, miR-223, miR-422a, miR-491-5p, miR-584, miR-664 and miR-1275 were significantly up-regulated, whereas only miR-20b was down-regulated [11].
Real-time quantitative PCR analysis showed that miR-145 expression increased dramatically in PBMCs from patients with MS [12]. An earlier study discovered that miR-142-3p, miR-146a, miR-155 and miR-326 were up-regulated in the PBMCs of MS patients [13]. Other studies showed that miR-21, miR-146a, miR-146b and miR-326 were up-regulated in PBMCs of RRMS patients as compared with controls [14, 15]. Analysis by using a microbead-based technology showed that miR-19a, miR-92a, miR-155 and let-7f were significantly up-regulated in the PBMCs of MS patients [16]. In comparison with controls, miRNA microarray analysis revealed that 45 miRNAs including miR-223*, miR-524-3p and miR-550* were increased, whereas 59 miRNAs including miR-31*, miR-150, miR-181c, miR-363, miR-374a*, miR-876-3p and let-7g were decreased in PBMCs from MS patients [17]. Another study discovered that the expression of miR-32, miR-125a, miR-146b, miR-200c, miR-210, miR-505, miR-582, miR-629 and miR-1180 increased significantly in the PBMCs from patients with MS, whereas the expression of miR-152, miR-197, miR-199a, miR-328, miR-363 and let-7g-3p decreased dramatically as compared with health controls in Chinese population [18].
miRNAs expressed differently in T Cells including CD4+ and CD8+ T cells
Microarray analysis revealed that the expression of 21 miRNAs including miR-15b, miR-23a, miR-30c, miR-99b, miR-125a-5p, miR-150, miR-197, miR-320d, miR-339-5p, miR-361-5p, miR-423-3p, miR-423-5p, miR-494, miR-625, miR-663, miR-1260b, miR-1280, miR-1979, miR-3153, miR-3178 and let-7a were significantly down-regulated in T cells from peripheral blood samples of MS patients [19].
It has been shown that both miR-15a and miR-16-1 were down-regulated in CD4+ T cells from peripheral blood samples of RRMS patients [20]. miR-27b, miR-128 and miR-340, however, were up-regulated in CD4+ T cells from patients with MS [21]. TaqMan array analysis indicated that 9 miRNAs including miR-1, miR-17-5p, miR-126, miR-193a, miR-200b, miR-376a, miR-485-3p, miR-486 and miR-497 were significantly increased, whereas miR-34a was decreased in CD4+ T cells from PBMCs of RRMS patients [22]. Further studies discovered that 17 miRNAs including miR-15a, miR-19a, miR-19b, miR-22, miR-29a, miR-29c, miR-93, miR-106b, miR-107, miR-148a, miR-210, miR-221, miR-223, miR-301a, miR-489, miR590-5p and let-7i were significantly up-regulated, whereas 7 miRNAs including miR-138-2*, miR-324-3p, miR-338-5p, miR-489, miR-512-3p, miR-564 and miR-886-3p were significantly down-regulated in regulatory T cells (Tregs) from peripheral blood samples of MS patients [23].
It has been reported that miR-629 was up-regulated, but miR-30a-3p, miR-149 and miR-497 were down-regulated in CD8+ T cells from peripheral blood samples of RRMS patients [22].
Unusual expression of miRNAs in B Cells and monocytes
TaqMan array analysis showed that miR-497 was up-regulated, but miR-92, miR-135b, miR-153, miR-189 and miR-422a were significantly down-regulated in B cells from peripheral blood samples of RRMS patients [22]. Microarray analysis discovered that the expression of 49 miRNAs including miR-7-1*, miR-15a, miR-16, miR-19b, miR-25, miR-28-5p, miR-93, miR-103-2, miR-106b, miR-107, miR-130b, miR-140-5p, miR-151-5p, miR-152, miR-181a, miR-191, miR-200a, miR-200b, miR-203, miR-204, miR-218, miR-221, miR-297a, miR-299a-5p, miR-320b, miR-329, miR-337-3p, miR-340*, miR-363, miR-369-5p, miR-411*, miR-486-5p, miR-515, miR-520a-3p, miR-520g, miR-548c-5p, miR-551a, miR-582-5p, miR-585, miR-599, miR-616*, miR-624*, miR-644, miR-649, miR-655 and let-7i were significantly down-regulated, but no miRNAs were found to be significantly up-regulated in the B lymphocytes from peripheral blood samples of RRMS patients [24]. Some of the down-regulated miRNAs may target the genes encoding the B cell receptor, phosphatidyl-inositol-3-kinase (PI3K) and phosphatase and tensin homology (PTEN) signaling pathways [24].
In one study, miR-155 expression was shown to be significantly increased in peripheral circulating CD14+ monocytes from patients with MS as compared with monocytes from control donors [25].
Dysregulation of miRNAs in Brain
It has been shown that miR-155 expression significantly increased in active brain lesions from patients with MS as compared with control donors [25-27]. miR-326 was also up-regulated in active MS lesions [27]. Quantitative real-time PCR analysis revealed that 20 miRNAs including miR-15a, miR-21, miR-22, miR-23a, miR-27a, miR-34a, miR-130a, miR-142-3p, miR-142-5p, miRNA-146a, miR-146b, miR-155, miR-193a, miR-199a, miR-200c, miRNA-214, miR-223, miR-320, miR-326 and miR-650 were up-regulated in the active brain white matter lesions from patients with MS, whereas 8 miRNAs including miR-23b, miR-139, miR-181c, miR-184, miR-328, miR-340, miR-487b and miR-656 were down-regulated as compared with the normal brain white matter tissues [28]. An earlier study discovered that the expression of miR-155, miR-338 and miR-491 increased in the cerebral white matter from MS patients as compared with patients without MS [29]. In addition, the expression of miR-124 increased in the brain hippocampi from patients with MS [30, 31].
Studies showed that miR-181c and miR-633 were up-regulated, whereas miR-922 was down-regulated in the cerebrospinal fluid of MS patients as compared with other neurologic diseases [32].
Abnormal expression of miRNAs in Experimental Autoimmune Encephalomyelitis (EAE)
EAE is a mouse model for MS. Real-time quantitative PCR analysis indicated that miRNA-21, miRNA-142-3p, miRNA-146a, miRNA-146b and miRNA-155 were up-regulated in the spinal cords of EAE mice and marmoset EAE brains [33]. miRNA-155 expression has also been shown to increase significantly in the spleen, lymph node and brain of EAE mice [9, 26]. Expression of let-7e also increased in CD4+ T cells and infiltrated mononuclear cells of brain and spinal cord from EAE mice [34]. miR-326 expression was significantly increased in peripheral blood leukocytes and CD4+ T cells from EAE mice [14]. miR-23b expression, however, decreased significantly in the spinal cords of EAE mice [35].
miRNA Signature in MS
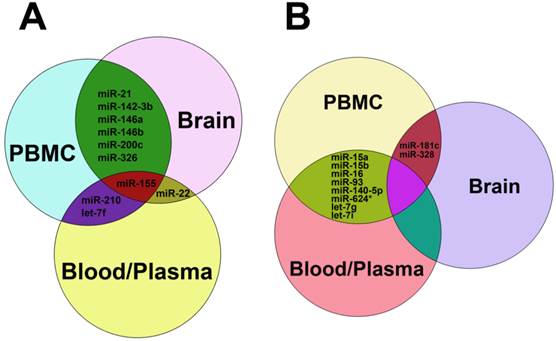
As stated above, miRNAs were highly dysregulated in MS patients and EAE mouse model. We summarized the up-regulated miRNAs (Table 1) and down-regulated miRNAs (Table 2) in different samples from MS patients and EAE mice. It is clear that miR-15a, miR-19a, miR-22, miR-210 and miR-223 were up-regulated in both Treg cells and other samples such as plasma, blood cells, PBMCs and brain white matter tissues from MS patients (Table 1), suggesting that these up-regulated miRNAs and Treg cells may play a role in MS pathogenesis. Besides, miR-21, miR-142-3p, miR-146a, miR-146b, miR-155 and miR-326 were up-regulated in both PBMCs and brain white matter lesions from MS patients and EAE mice (Table 1 and Fig. 3A), suggesting that these up-regulated miRNAs may be used as a signature for MS and play critical roles in MS pathogenesis. Similarly, miR-181c and miR-328 were down-regulated in both PBMCs and brain white matter lesions, and miR-15a and miR-15b were down-regulated in blood, peripheral T cells and B cells or plasma samples from MS patients (Table 2 and Fig. 3B), suggesting that these down-regulated miRNAs have implications in MS pathogenesis. Notably, miR-15a was up-regulated in Treg cells and brain white matter lesions (Table 1), but down-regulated in blood, peripheral T cells and B cells (Table 2), suggesting that regulation of miRNA expression in MS was complicated.
Comparison of up-regulated miRNAs expressed in different samples from MS patients and EAE mice
| Sample | Blood | Plasma | Blood cells | PBMCs | T cells# | Tregs | B cells | Brain lesions | EAE mice |
|---|---|---|---|---|---|---|---|---|---|
| Number of miRNAs analyzed | 2 | 35 | 9 | 21 | 13 | 17 | 1 | 22 | 7 |
| miR-15a | miR-15a | ||||||||
| miR-19a | miR-19a | ||||||||
| miR-21 | miR-21 | miR-21 | |||||||
| miR-22 | miR-22 | miR-22 | |||||||
| miR-142-3p | miR-142-3p | miR-142-3p | |||||||
| miR-145 | miR-145 | ||||||||
| miR-146a | miR-146a | miR-146a | |||||||
| miR-146b | miR-146b | miR-146b | |||||||
| miR-155 | miR-155 | miR-155 | miR-155 | ||||||
| miR-193a | miR-193a | ||||||||
| miR-200c | miR-200c | ||||||||
| miR-210 | miR-210 | miR-210 | |||||||
| miR-223 | miR-223 | miR-223 | |||||||
| miR-326 | miR-326 | miR-326 | |||||||
| miR-497 | miR-497 | ||||||||
| miR-629 | miR-629 | ||||||||
| let-7f | let-7f | ||||||||
| Reference | [5, 6] | [7-9] | [11] | [12-18] | [21, 22] | [23] | [22] | [25-29] | [9, 14, 26, 33, 34] |
# T cells were total T cells including T cells, CD4+ and CD8+ T cells.
miRNAs in bold were those expressed in Treg cells and other samples including brain, blood cells, PBMC and plasma.
miRNAs in bold and italics were those expressed in PBMCs and other samples including brain, plasma and EAE mice.
Comparison of down-regulated miRNAs expressed in different samples from MS patients and EAE mice
| Sample | Blood | Plasma | Blood cells | PBMCs | T cells# | Tregs | B cells | Brain | EAE mice |
|---|---|---|---|---|---|---|---|---|---|
| Number of miRNAs analyzed | 27 | 4 | 1 | 12 | 27 | 7 | 51 | 8 | 1 |
| miR-15a | miR-15a | miR-15a | |||||||
| miR-15b | miR-15b | miR-15b | |||||||
| miR-16 | miR-16 | ||||||||
| miR-20b | miR-20b | ||||||||
| miR-23a | miR-23a | ||||||||
| miR-93 | miR-93 | ||||||||
| miR-140-5p | miR-140-5p | ||||||||
| miR-150 | miR-150 | ||||||||
| miR-152 | miR-152 | ||||||||
| miR-181c | miR-181c | ||||||||
| miR-197 | miR-197 | ||||||||
| miR-328 | miR-328 | ||||||||
| miR-363 | miR-363 | ||||||||
| miR-624* | miR-624* | ||||||||
| miR-1979 | miR-1979 | ||||||||
| let-7g | let-7g | ||||||||
| let-7i | let-7i | ||||||||
| Reference | [5, 6] | [8, 10] | [11] | [17, 18] | [19, 20, 22] | [23] | [22, 24] | [28] | [35] |
# T cells were total T cells including T cells, CD4+ and CD8+ T cells.
miRNAs in bold were those expressed in PBMCs and brain lesions of MS patients.
miRNAs in bold and italics were those expressed in three different samples including blood, B cells and T cells or blood, plasma and T cells.
Regulation of miRNAs in MS
Interferon-beta (IFN-β) is broadly used in the treatment of patients with MS [36]. It has been reported that 7 miRNAs, including miR-16-5p, miR-342-5p, miR-346, miR-518b, miR-760, let-7a-5p and let-7b-5p, were increased, whereas 13 miRNAs, including miR-27a-5p, miR-29a-3p, miR-29b-1-5p, miR-29c-3p, miR-95, miR-149-5p, miR-181c-3p, miR-193a-3p, miR-193-5p, miR-423-5p, miR-532-5p, miR-708-5p and miR-874, were decreased in PBMCs from MS patients after IFN-β therapy [37], which were possibly related with apoptotic processes and IFN feedback loops. In addition, IL-17 was usually up-regulated in MS patients and EAE mice [38]. Studies showed that IL-17 suppressed miR-23b expression, which was responsible for the decrease of miR-23b expression in the spinal cords of EAE mice, and in turn, miR-23b down-regulated IL-17 [35]. The results suggested that cytokines may regulate miRNA expression in MS.
Glatiramer acetate (GA), an immunomodulation drug, is currently used to treat MS [39, 40]. GA treatment induced significant decreases of miR-146a and miR-142-3p expression in the PBMCs of MS patients [13]. Since miR-146a and miR-142-3p were up-regulated in PBMCs, brain white matter lesions of MS patients and EAE mice (Table 1), GA may regulate miRNA expression and thus improve disease status of MS patients.
Natalizumab is a monoclonal antibody against α4-integrin and is used in the treatment of MS patients [41]. miRNA microarray analysis found that 10 miRNAs including miR-19b, miR-106b, miR-142-5p, miR-150, miR-191, miR-383, miR-551a and miR-598 were up-regulated in B lymphocytes of peripheral blood samples from RRMS patients 6 months after natalizumab treatment as compared with untreated RRMS patients [24]. In addition, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), an activator for aryl hydrocarbon receptor, could induce anti-inflammation and increase miR-132 expression in EAE mice, and thus suppress EAE [42]. The results suggest that non-immune modulated drugs may also regulate miRNA expression in MS and could potentially be used in the treatment of MS patients.
Function of miRNAs in MS
As indicated above, while most studies have shown altered expression of miRNA during MS, the significance of this is unclear. It has been shown that miRNAs are involved in several crucial biological functions and human diseases such as neurological disorders [3]. Thus, it is not surprising that dysregulated miRNAs play an important role in MS and EAE development. It has been reported that miR-18b and miR-599 may play a role at the time of relapse and miR-96 may be associated with remission of RRMS patients [43].
It has been documented that miR-155 could induce the development of Th1 and Th17 cells, leading to severe disease in both patients with MS and mice with EAE [9]. Similarly, deficiency in the miR-132/212 cluster induced the lower frequencies of Th1 and Th17 cells and resistance to EAE development in mice [44]. Furthermore, let-7e knock-down attenuated Th1 and Th17 cells [34] and in vivo silencing of miR-326 suppressed Th17 cell development [14], leading to the inhibition of EAE. It has also been reported that miR-23b could inhibit IL-17 expression, which plays an important role in autoimmune pathogenesis [35]. The results suggest that miRNAs in the regulation of Th1 cells, IL-17 and Th17 cells may have great implications in the pathogenesis of MS.
Studies discovered that the expression of miR-27b, miR-128 and miR-340 significantly increased in CD4+ T cells from patients with MS [21]. Further studies found that miR-27b, miR-128 and miR-340 inhibited expression of B lymphoma Mo-MLV insertion region 1 homolog (BMI1) and IL-4, leading to decreased levels of GATA3 and a shift from Th2 to Th1 cytokines [21]. Therefore, miR-27b, miR-128 and miR-340 may play an important role in MS pathogenesis.
It has been shown that the up-regulation of miR-124 in the hippocampi was associated with demyelination in the brain of patients with MS [30]. Other studies revealed that miR-34a, miR-155 and miR-326 increased in active multiple sclerosis lesions and decreased CD47 expression, leading to macrophage release from inhibitory control and phagocytosis of myelin [28]. The results suggest that specific miRNAs may have a critical role in demyelination, which contributes to MS pathogenesis.
The dysfunction of the blood-brain barrier (BBB) was reported to be associated with the pathogenesis of MS and down-regulation of miR-125a-5p was involved in the regulation of immune cell efflux and tightness of endothelial cells in the BBB [45], suggesting that miR-125a-5p may play a role in MS.
Database analysis on miRNA-mRNA interactions showed that MS-associated miR-20a-5p and miR-20b-5p occurred as central hubs in the regulation of one thousand targeted genes [46], suggesting that miR-20a-5p and miR-20b-5p may play an important role in MS pathogenesis.
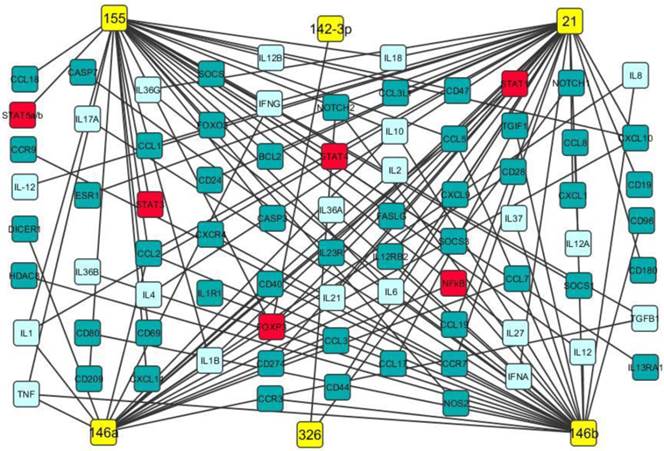
Our data analysis discovered that miR-21, miR-142-3p, miR-146a, miR-146b, miR-155 and miR-326 were up-regulated in both PBMCs and brain white matter lesions from MS patients and EAE mice as well (Table 1). It has been documented that miR-21 promoted IFN-γ and IL-17A production in T cells, which were responsible for Th1 and Th17 development, targeted programmed cell death protein 4 (PDCD4) and thus inhibited apoptosis [47]. miR-142-3p not only inhibited IL-10 expression in CD4+ T cells [48], but also targeted D1 dopamine receptor in the brain and suppressed neuronal growth and development [49]. miR-146a was positively correlated with IL-17 production by inhibiting target gene IRAK1 [50] and regulated inflammation in human neurological disorders by targeting IL-6 and COX-2 [51]. miR-146b may have anti-inflammatory activity by activating the NF-κB pathway [52] and inhibiting proinflammatory cytokine and chemokine production [53]. It has been demonstrated that miR-155 promotes Th1 and Th17 cells and miR-326 increases Th17 cells, leading to severe disease in MS patients and EAE mice [9, 14]. Potential gene targets of miR-21, miR-142-3p, miR-146a, miR-146b, miR-155 and miR-326 include transcription factors (such as STAT1, STAT4, and FoxP3), chemokines, cytokines (such as IFN-γ, TNF-α, TGF-β, IL-2, IL-6 and IL-17), cell apoptosis regulators and growth factors (Fig. 2). Therefore, these up-regulated miRNAs may play critical roles in MS pathogenesis (Fig. 2 and Table 3).
Potential gene targets of miR-21, miR-142-3p, miR-146a, miR-146b, miR-155 and miR-326. Potential gene targets including cytokine (light blue boxes), transcription factor (red boxes) and chemokine of miR-21, miR-142-3p, miR-146a, miR-146b, miR-155 and miR-326 (yellow boxes) were predicted by Ingenuity IPA online analysis and the figure was created by Cytoscape 3.0.1 software.
Functions of dysregulated miRNAs in patients with multiple sclerosis
| miRNA | Expression | Function | Biomarker | Therapeutic target | Reference |
|---|---|---|---|---|---|
| miR-7-1-3p | Down-regulation | Blood | [5] | ||
| miR-15b | Down-regulation | Serum | [10] | ||
| miR-20a-5p | Down-regulation | Blood | [5] | ||
| miR-21 | Up-regulation | Th1 and Th17 increase | [14, 15] | ||
| miR-23a | Down-regulation | Serum | [10] | ||
| miR-23b | Up-regulation | Th17 increase | Potential | [35] | |
| miR-27b | Up-regulation | Th1 increase | Potential | [21] | |
| miR-92a-1* | Up-regulation | Plasma | [7] | ||
| miR-124 | Up-regulation | Potential | [30, 31] | ||
| miR-125a-5p | Down-regulation | Potential | [45] | ||
| miR-128 | Up-regulation | Th1 increase | Potential | [21] | |
| miR-145 | Up-regulation | Plasma, PBMC | [7, 12] | ||
| miR-146a | Up-regulation | Th17 increase | [13-15] | ||
| miR-155 | Up-regulation | Th1 and Th17 increase | Plasma, PBMC, Brain | Potential | [9, 13, 25-27] |
| miR-223 | Down-regulation | Serum | [10] | ||
| miR-326 | Up-regulation | Th17 increase | Potential | [13-15, 27] | |
| miR-340 | Up-regulation | Th1 increase | Potential | [21] | |
| let-7e | Up-regulation | Th1 and Th17 increase | [7] |
miRNAs as MS Diagnostic Markers
Results have demonstrated that miR-155 expression was significantly increased in both patients with MS and mice with EAE [9, 26], suggesting that miR-155 may be used as a marker for patients with MS. Since miR-145 increased dramatically in PBMCs from patients with multiple sclerosis [12] and could be used in the discrimination of MS patients from healthy controls [11], miR-145 may be used as a good diagnostic biomarker for multiple sclerosis.
It has been shown that the expression of miR-20a-5p and miR-7-1-3p decreased in the blood of patients with MS and thus miR-20a-5p and the other validated miRNAs may be used as biomarkers for MS [5]. miR-92a-1*, miR-145 and let-7 family of miRNAs increased significantly in the plasma of patients with MS [7], whereas miR-15b, miR-23a and miR-223 were down-regulated in the serum samples from patients with MS as compared with normal controls [10]. The results suggest that circulating miRNAs may be used as blood biomarkers to diagnose MS.
Our data analysis demonstrated that miR-21, miR-142-3p, miR-146a, miR-146b, miR-155 and miR-326 were up-regulated in PBMCs and brain white matter lesions from MS patients (Table 1). Therefore, it is possible that these up-regulated miRNA may be used as diagnostic markers for MS (Table 3).
Common up-regulated (A) and down-regulated miRNAs (B) in blood, plasma, PBMC and brain tissues from patients with MS. In panel A, miRNAs shown in the green zone were up-regulated in both PBMC and brain, those in the purple zone were up-regulated in both PBMC and blood/plasma, miR-22 was up-regulated in both brain and blood/plasma, and miR-155 was up-regulated among PBMC, brain and blood/plasma. In panel B, miRNAs in the reseda green zone were down-regulated in both PBMC and blood/plasma, and miR-181c and miR-328 were down-regulated in both brain and blood/plasma.
miRNAs as MS Therapeutic Targets
Studies have demonstrated that suppression of miR-155 expression could inhibit the development of Th1 and Th17 cells, resulting in the decrease of disease severity in mice with EAE [9]. Furthermore, anti-miR-155 treatment significantly inhibited EAE development [54]. It has been demonstrated that miR-155 expression increased significantly in patients with MS [25-27].Therefore, miR-155 may be a good therapeutic target for MS.
It appeared that miR-326 expression increased significantly in active MS lesions, and in vivo silencing of miR-326 significantly inhibited Th17 cells and dramatically decreased EAE severity [14, 27]. The results suggest that miR-326 may be a therapeutic target for MS.
It has been reported that miR-23b down-regulated IL-17, inhibited tumor necrosis factor alpha (TNF-α)- or IL-1beta-induced nuclear factor kappa-B (NF-κB) activation, and suppressed TGF-β-activated kinase 1/MAP3K7 binding protein 2 (TAB2), TAB3 and NF-κB kinase subunit alpha (IKK-α) in EAE mice [35], suggesting that miR-23b could be used in the treatment of MS.
It has been shown that down-regulation of miR-125a-5p was responsible for the BBB dysregulation associated with MS, and thus miR-125a-5p may be used in the recovery of BBB function for effective treatment of MS [45].
Studies showed that miR-124 expression increased in the demyelinated brain hippocampi from patients with MS [30, 31], suggesting that miR-124 may play a role in the pathogenesis of MS and could be a therapeutic target for MS patients.
It has been reported that miR-27b, miR-128 and miR-340 were up-regulated in CD4+ T cells from patients with MS and induced a shift from Th2 to Th1 cytokines [21]. In vitro treatment of oligonucleotide micro-RNA inhibitors for miR-27b, miR-128 and miR-340 induced the restoration of Th2 responses in T cells from MS patients [21]. The results suggest that in vivo silencing of miR-27b, miR-128 and miR-340 may potentially be used in the treatment of MS patients.
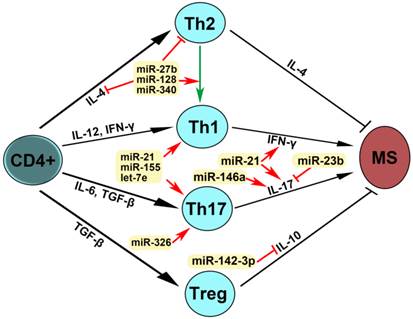
miRNAs were firstly identified in the early 1990s [55]. Recently, studies have demonstrated that miRNAs have multiple functions in negative gene regulation and play important roles in most biological processes [56-58]. In patients with MS, reports have shown that miR-27b, miR-128 and miR-340 inhibit IL-4 expression, suppress Th2 development and induce a shift from Th2 to Th1, leading to MS pathogenesis (Fig. 4). miR-21, miR-155 and let-7e promote Th1 and Th17 development, miRNA-21 increases the expression of both IFN-γand IL-17, miR-146a increases IL-17 expression, miRN-326 promotes Th17 development, and miR-142-3p inhibits IL-10 expression, which contribute to MS pathogenesis. miR-23b suppressed IL-17 expression and thus inhibited MS pathogenesis. These dysregulated miRNAs may be used as therapeutic targets for MS (Fig. 4). Therefore, studies on the expression, regulation and function of miRNAs in patients with multiple sclerosis will provide the unique insights for the development of specific miRNAs as diagnostic markers and therapeutic targets for multiple sclerosis in the future (Table 3, Figs. 3 and 4).
Mechanism of action of dysregulated miRNAs in patients with MS. IL-4 is required for the differentiation of naïve CD4+ T cells into Th2. IL-12 and IFN-γ promoted Th1 differentiation. Il-6 and TGF-β stimulated Th17 differentiation. TGF-β induced Treg differentiation. Th2 produces IL-4 and Treg secrets IL-10, leading to the inhibition of MS, whereas Th1 expresses IFN-γ and Th17 produces IL-17, which induce MS pathogenesis. An arrow represents promotion and a T shaped line represents inhibition. IL, interleukin; IFN-γ, interferon-gamma; TGF-β, transforming growth factor-beta.
Abbreviations
miRNA: microRNA; MS: multiple sclerosis; RR: relapsing-remitting; EAE: experimental autoimmune encephalomyelitis; PBMC: peripheral blood mononuclear cell; Treg: regulatory T cell; PCR: polymerase chain reaction; RT-PCR: reverse transcription-PCR
Acknowledgements
This work was supported by a grant (#81341037 to JZ) from the National Natural Science Foundation of China and “Taishan Scholar” Special Fund (#109, 2012 to JZ) from the Shandong Government, China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Compston A, Coles A. Multiple sclerosis. Lancet. 2002;359:1221-31
2. Milo R, Kahana E. Multiple sclerosis: geoepidemiology, genetics and the environment. Autoimmun Rev. 2010;9:A387-94
3. Tarassishin L, Loudig O, Bauman A. et al. Interferon regulatory factor 3 inhibits astrocyte inflammatory gene expression through suppression of the proinflammatory miR-155 and miR-155*. Glia. 2011;59:1911-22
4. Smith KM, Guerau-de-Arellano M, Costinean S. et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189:1567-76
5. Keller A, Leidinger P, Steinmeyer F. et al. Comprehensive analysis of microRNA profiles in multiple sclerosis including next-generation sequencing. Mult Scler. 2014;20:295-303
6. Cox MB, Cairns MJ, Gandhi KS. et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PloS One. 2010;5:e12132
7. Gandhi R, Healy B, Gholipour T. et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73:729-40
8. Siegel SR, Mackenzie J, Chaplin G. et al. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep. 2012;39:6219-25
9. Zhang J, Cheng Y, Cui W. et al. MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. J Neuroimmunol. 2013;266:56-63
10. Fenoglio C, Ridolfi E, Cantoni C. et al. Decreased circulating miRNA levels in patients with primary progressive multiple sclerosis. Mult Scler. 2013;19:1938-42
11. Keller A, Leidinger P, Lange J. et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PloS One. 2009;4:e7440
12. Sondergaard HB, Hesse D, Krakauer M. et al. Differential microRNA expression in blood in multiple sclerosis. Mult Scler. 2013;19:1849-57
13. Waschbisch A, Atiya M, Linker RA. et al. Glatiramer acetate treatment normalizes deregulated microRNA expression in relapsing remitting multiple sclerosis. PloS One. 2011;6:e24604
14. Du C, Liu C, Kang J. et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252-9
15. Fenoglio C, Cantoni C, De Riz M. et al. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci Lett. 2011;504:9-12
16. Paraboschi EM, Solda G, Gemmati D. et al. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int J Mol Sci. 2011;12:8695-712
17. Martinelli-Boneschi F, Fenoglio C, Brambilla P. et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci Lett. 2012;508:4-8
18. Yang D, Wang WZ, Zhang XM. et al. MicroRNA Expression Aberration in Chinese Patients with Relapsing Remitting Multiple Sclerosis. J Mol Neurosci. 2014;52:131-7
19. Jernas M, Malmestrom C, Axelsson M. et al. MicroRNA regulate immune pathways in T-cells in multiple sclerosis (MS). BMC Immunol. 2013;14:32
20. Lorenzi JC, Brum DG, Zanette DL. et al. miR-15a and 16-1 are downregulated in CD4+ T cells of multiple sclerosis relapsing patients. Int J Neurosci. 2012;122:466-71
21. Guerau-de-Arellano M, Smith KM, Godlewski J. et al. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain. 2011;134:3578-89
22. Lindberg RL, Hoffmann F, Mehling M. et al. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol. 2010;40:888-98
23. De Santis G, Ferracin M, Biondani A. et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol. 2010;226:165-71
24. Sievers C, Meira M, Hoffmann F. et al. Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin Immunol. 2012;144:70-9
25. Moore CS, Rao VT, Durafourt BA. et al. miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann Neurol. 2013;74:709-20
26. Thamilarasan M, Koczan D, Hecker M. et al. MicroRNAs in multiple sclerosis and experimental autoimmune encephalomyelitis. Autoimmun Rev. 2012;11:174-9
27. Junker A. Pathophysiology of translational regulation by microRNAs in multiple sclerosis. FEBS Lett. 2011;585:3738-46
28. Junker A, Krumbholz M, Eisele S. et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342-52
29. Noorbakhsh F, Ellestad KK, Maingat F. et al. Impaired neurosteroid synthesis in multiple sclerosis. Brain. 2011;134:2703-21
30. Shapshak P. Molecule of the month: miRNA and Multiple Sclerosis. Bioinformation. 2013;9:847-8
31. Dutta R, Chomyk AM, Chang A. et al. Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microRNA miR-124 and reduced AMPA receptors. Ann Neurol. 2013;73:637-45
32. Haghikia A, Hellwig K, Baraniskin A. et al. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology. 2012;79:2166-70
33. Lescher J, Paap F, Schultz V. et al. MicroRNA regulation in experimental autoimmune encephalomyelitis in mice and marmosets resembles regulation in human multiple sclerosis lesions. J Neuroimmunol. 2012;246:27-33
34. Guan H, Fan D, Mrelashvili D. et al. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur JImmunol. 2013;43:104-14
35. Zhu S, Pan W, Song X. et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-alpha. Nat Med. 2012;18:1077-86
36. Kremenchutzky M, Morrow S, Rush C. The safety and efficacy of IFN-beta products for the treatment of multiple sclerosis. Expert Opin Drug Saf. 2007;6:279-88
37. Hecker M, Thamilarasan M, Koczan D. et al. MicroRNA expression changes during interferon-beta treatment in the peripheral blood of multiple sclerosis patients. Int J Mol Sci. 2013;14:16087-110
38. Pollinger B. IL-17 producing T cells in mouse models of multiple sclerosis and rheumatoid arthritis. J Mol Med. 2012;90:613-24
39. Messina S, Patti F. The pharmacokinetics of glatiramer acetate for multiple sclerosis treatment. Expert OpinDrug Metab Toxicol. 2013;9:1349-59
40. Johnson KP, Due DL. Benefits of glatiramer acetate in the treatment of relapsing-remitting multiple sclerosis. Expert RevPharmacoecon Outcomes Res. 2009;9:205-14
41. Brown BA. Natalizumab in the treatment of multiple sclerosis. Ther Clin Risk Manag. 2009;5:585-94
42. Hanieh H, Alzahrani A. MicroRNA-132 suppresses autoimmune encephalomyelitis by inducing cholinergic anti-inflammation: a new Ahr-based exploration. Eur J Immunol. 2013;43:2771-82
43. Otaegui D, Baranzini SE, Armananzas R. et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PloS One. 2009;4:e6309
44. Nakahama T, Hanieh H, Nguyen NT. et al. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc Natl Acad Sci U S A. 2013;110:11964-9
45. Reijerkerk A, Lopez-Ramirez MA, van Het Hof B. et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: implications for multiple sclerosis. J Neurosci. 2013;33:6857-63
46. Angerstein C, Hecker M, Paap BK. et al. Integration of MicroRNA databases to study MicroRNAs associated with multiple sclerosis. Mol Neurobiol. 2012;45:520-35
47. Ando Y, Yang GX, Kenny TP. et al. Overexpression of microRNA-21 is associated with elevated pro-inflammatory cytokines in dominant-negative TGF-beta receptor type II mouse. J Autoimmun. 2013;41:111-9
48. Ding S, Liang Y, Zhao M. et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis and rheum. 2012;64:2953-63
49. Tobon KE, Chang D, Kuzhikandathil EV. MicroRNA 142-3p mediates post-transcriptional regulation of D1 dopamine receptor expression. PloS One. 2012;7:e49288
50. Xia P, Fang X, Zhang ZH. et al. Dysregulation of miRNA146a versus IRAK1 induces IL-17 persistence in the psoriatic skin lesions. Immunol Lett. 2012;148:151-62
51. Iyer A, Zurolo E, Prabowo A. et al. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PloS One. 2012;7:e44789
52. Nata T, Fujiya M, Ueno N. et al. MicroRNA-146b improves intestinal injury in mouse colitis by activating nuclear factor-kappaB and improving epithelial barrier function. J Gene Med. 2013;15:249-60
53. Curtale G, Mirolo M, Renzi TA. et al. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci U S A. 2013;110:11499-504
54. Murugaiyan G, Beynon V, Mittal A. et al. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213-21
55. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-54
56. Arora S, Rana R, Chhabra A. et al. miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288:77-87
57. Singh RP, Massachi I, Manickavel S. et al. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12:1160-5
58. Bronevetsky Y, Ansel KM. Regulation of miRNA biogenesis and turnover in the immune system. Immunol Rev. 2013;253:304-16
Author contact
![]() Corresponding author: J.-H. Zhou, PhD, Institute for Tumor Immunology, Ludong University School of Life Sciences, 186 Hongqi Middle Road, Yantai, Shandong 264025, PR China, Tel: 86-183-0050-1903; Fax: 86-0535-664-2910; E-mail: Juhua.zhoucom
Corresponding author: J.-H. Zhou, PhD, Institute for Tumor Immunology, Ludong University School of Life Sciences, 186 Hongqi Middle Road, Yantai, Shandong 264025, PR China, Tel: 86-183-0050-1903; Fax: 86-0535-664-2910; E-mail: Juhua.zhoucom

 Global reach, higher impact
Global reach, higher impact