Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(5):404-408. doi:10.7150/ijms.8033 This issue Cite
Short Research Communication
All Human EF1α Promoters Are Not Equal: Markedly Affect Gene Expression in Constructs from Different Sources
Molecular Physiology and Therapeutics Branch, National Institute of Dental and Craniofacial Research, NIH, DHHS, Bethesda, MD 20892-1190.
Received 2013-11-1; Accepted 2014-1-9; Published 2014-3-7
Abstract
The promoter is a major element in the expression cassette of gene therapy vectors. Optimal promoter selection can enhance target specificity and gene expression. Recently, we evaluated three different human elongation factor 1 alpha (EF1α) promoters. The three promoters were put into the same expression vector, pAC-luc, driving expression of the luciferase cDNA. The activity from one EF1α promoter (termed EF1α -3), obtained in a commercial vector, was markedly lower when tested in vitro (from 50 - 500 x) in four cell lines and in vivo in rat submandibular glands (~250 x). Sequence differences in the EF1α -3 promoter likely account for the activity differences seen. Investigators need to recognize that all promoters of the same name may not be equivalent in driving transgene expression.
Keywords: promoter, EF1α, gene therapy.
Introduction
The expression cassette is an essential component of a gene therapy vector, and each includes at least three key components: a promoter, a transgene and a polyadenylation sequence. The optimal design of an expression cassette can facilitate therapeutic outcomes (1, 2, 3). The promoter plays a critical role, including directing, regulating and targeting transgene expression. Higher promoter activity can result in lower doses of vector administered, with consequently lower toxicity or side effects.
Currently, many promoters are available for the gene therapy studies. Differences, however, are known to exist within promoters of the same name. For example, the CMV promoter (including human cytomegalovirus enhancer and immediate early promoter) used in different vectors was 589 bp in size in Ad-EGFP (4), 646 bp in pCNS (5), 742 bp in phMGFP (accession No. in NCBI: AY218848), 795 bp in pCMVTNT(TM)(accession No. in NCBI: AF477200), 817 bp in expression vector M64754 (accession No. in NCBI: M64754) and 1650 bp in expression vector AF286076 (accession No. in NCBI: AF286076). All are simply called the CMV promoter. It is not known whether such sequence differences result in significant functional differences.
Recently, in an effort to optimize expression cassettes for planned human trials, we have studied three versions of the human EF1α promoter, two obtained from commercial vectors and one from a colleague. Although, all are called the EF1α promoter, the functional activity from one commercially-obtained promoter was markedly lower in four different cell lines in vitro and in vivo in rat submandibular glands.
Materials and Methods
Construction of plasmids
A backbone plasmid, pAC-luc (6), was constructed using pACCMV-pLpA that was a gift from Dr. C. Newgard (University of Texas, Southwestern Medical Center). This plasmid contains part of the adenovirus serotype 5 genome (0-1.2 and 9.2-17 mu) with the human CMV immediate early promoter/enhancer substituted between map units 1.3 and 9.1. pACCMV-pLpA was digested with Not I to delete the CMV promoter/enhancer and SV40 poly A sequence. A 307 bp DNA fragment, containing a multiple cloning site (Bgl II, Xho I, Hind III, EcoR I, Sal I, Kpn I, BamH I and Xba I) and SV40 poly A sequence, was removed from pEGFP-C3 (BD Biosciences, Palo Alto, CA) with Mlu I and Sca I, and filled in with Klenow large DNA fragment (Invitrogen, Carlsbad, CA). Not I linkers were added and then ligated with the remaining pACCMV-pLpA fragment to become the pAC plasmid. Next, a 1851 bp luciferase cDNA (Kpn I/BamH I fragment from pGL2-Basic, Promega, Madison, WI) was inserted into the pAC Kpn I/BamH I sites to generate pAC-luc. The first EF1α promoter used (1197 bp), termed EF1α-1 in this study, was excised from the plasmid pEAK8 (Edge BioSystems, Gaithersburg, MD) with Spe I, filled in with Klenow large DNA fragment, and then digested with Eco RI. Next, pAC-luc was digested with Hind III, filled in with Klenow large DNA fragment, then digested with Eco RI and ligated with EF1α-1. The second EF1α promoter, termed EF1α-2 (1272 bp), was a gift from Dr. S. Gutkind (OPCB, NIDCR, NIH) (7), and was excised from pEF-BOS with EcoR I/Hind III and inserted into pAC-luc. The last EF1α promoter studied here, termed EF1α-3 (1191 bp), was excised from pEF1/myc/ER/GFP (Invitrogen) with Eco RI/Nco I, filled in with Klenow large DNA fragment, and then Hind III linkers were added to both ends. EF1α-3 was inserted into pAC-luc at site the Hind III site.
Cell culture
The A5 cell line was derived from rat submandibular gland (8) and grown in McCoy's 5A medium (Invitrogen). The HSY cell line was obtained from a human parotid adenocarcinoma (9) and grown in a mixture of 50% Dulbecco's minimum essential medium (DMEM) and 50% Ham's F12 media. The HSG cell line was obtained from an irradiated human submandibular gland (10) and grown in DMEM/F12 medium, as above. The 293 cell line (Microbix Biosystems Inc., Toronto, Ontario, Canada) is from human embryonic kidney (11) and was grown in Eagle's minimum essential medium. For all four cell lines the following supplements were included: 10% fetal bovine serum (Invitrogen,), 100 U/ml penicillin G (Invitrogen), 100 µg/ml streptomycin (Invitrogen). Cells were incubated at 37ºC in a humidified 5% CO2 atmosphere.
In vitro and in vivo plasmid transfection
We used adenovirus- polyethylenimine (PEI)-plasmid complexes to deliver plasmids into A5, HSG, HSY and 293 cells in vitro and rat submandibular gland in vivo. PEI (Sigma, St. Louis, MO) has a high cationic-charge-density potential and is useful to enhance transfection (12,13). The adenovirus used for all transfections herein was Adcontrol, an E1- recombinant adenovirus without any transgene. Adcontrol was generated by cotransfection of pACCMV-pLpA and pJM17 in C7 cells (6). The complexes were formed using 2 x 1010 molecules of plasmid DNA/4 x 105 cells, 0.1 mM of PEI and 3 x 1010 particles of Adcontrol in a volume of 20 µl. A5, HSG, HSY and 293 cells were plated at 2 x 105 cells/well in 96 well plates. After 24 hours, the cells were transfected with 20 µl of adenovirus-PEI-plasmid complexes for 1 hour, then 180 µl of fresh growth medium was added to each well. Cells were cultured for an additional 24 hours and then luciferase activity was measured.
Male Wistar rats (250-350 g, ~3 months old) were anesthetized (14,15,16) and the adenovirus-PEI-plasmid complexes for plasmid transfection, which were formed using 4.35 x 1012 molecules of plasmid DNA/gland, 0.5 mM of PEI and 1 x 1011 particles of Adcontrol in a volume of 200 µl, were administered by retrograde ductal instillation (14,15,16) into both submandibular glands. Three rats were studied for each experimental group. After 3 days, rats were euthanized and submandibular glands collected for analysis of luciferase activity. All animal studies were approved by the NIDCR Animal Care and Use Committee.
Luciferase assays
A5, HSG, HSY and 293 cells were lysed 24 hours post-transfection with cell lysis buffer (Promega) for 15 min at room temperature. For rat submandibular tissues, about 100 mg wet weight of samples were added to 500 µl of 1 x cell lysis buffer (Promega). Samples were homogenized with a homogenizer from OMNI international (Waterbury, CT) and incubated for 15 min at room temperature. Fifty microliters of the cell lysates were transferred to assay tubes, 100 µl of luciferase substrate injected, and light output was measured with an OPTOCOMP I luminometer (GEM Instruments, INC., Hamden, CT) for a 10 sec interval. After the measurement of luciferase activity, the concentration of protein in separate aliquots of the lysate was measured using the BCA protein assay kit (PIERCE, Rockford, IL). Results were expressed as relative light unit (RLU)/ mg protein.
Data analysis
Data analyses employed SigmaStat version 2.0 (SPSS Inc, Chicago, IL). Descriptive statistics were calculated and are reported as mean ± SD. One-way ANOVAs, following by a Tukey test, were used to determine the statistical significance of differences. Values of p < 0.05 were considered significant.
Results and Discussion
EF1α promoter activity
On average, the EF1α-1 and EF1α-2 promoters showed the highest activities in all cell lines. The EF1α-1 promoter activity was statistically higher in A5 and HSG cells (~2 x; p< 0.001), but there were no statistical differences between the activity of these promoters in HSY and 293 cells (Figure 1). However, as seen in Figure 1, it is clear that luciferase activity with the EF1α-3 promoter was significantly lower than those seen with the EF1α-1 and EF1α-2 promoters in all four cell lines (p< 0.001); ~50-fold lower in A5 cells, ~145-fold lower in HSG cells, ~80-fold lower in HSY cells and ~500-fold lower in 293 cells.
Comparison of three EF1α promoter activities in vitro after transfection using plasmid-PEI-Adcontrol complexes. A. Promoter activity in A5 cells; B. Promoter activity in HSG cells; C. Promoter activity in HSY cells; D. Promoter activity in 293 cells. Each cell was transfected with 5 x 104 plasmid molecules. See text for details. The data shown are the means ± SD of luciferase activity from three determinations 24 hr after transfections and are representative of three separate experiments. For most determinations the error bars are too small to be visualized in the figure.

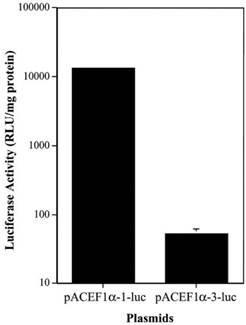
To determine if there was a similar difference in the activity of the different EF1α promoters in vivo, we tested two plasmids, pAC EF1α-1-luc and pAC EF1α-3-luc, in rat submandibular glands. The plasmids were transfected at 4.35 x 1012 molecules/gland using plasmid-PEI-Adcontrol complexes, and luciferase activity was measured at 3 days post-transfection (Figure 2). As the data in Figure 2 show, the luciferase activity observed in vivo with the EF1α-3 promoter was also markedly lower (~250-fold) than that with the EF1α-1 promoter (p< 0.01).
Comparison of two EF1α promoter activities in vivo in rat submandibular glands after transfection using plasmid-PEI-Adcontrol complexes. Each submandibular gland was transfected with 4.35 x 1012 plasmid molecules. See text for details. The data shown are the means ± SD of luciferase activity from six submandibular glands of three rats measured three days after transfections. Note that for the pAC EF1α-1-luc data the error bars are too small to be visualized in the figure.
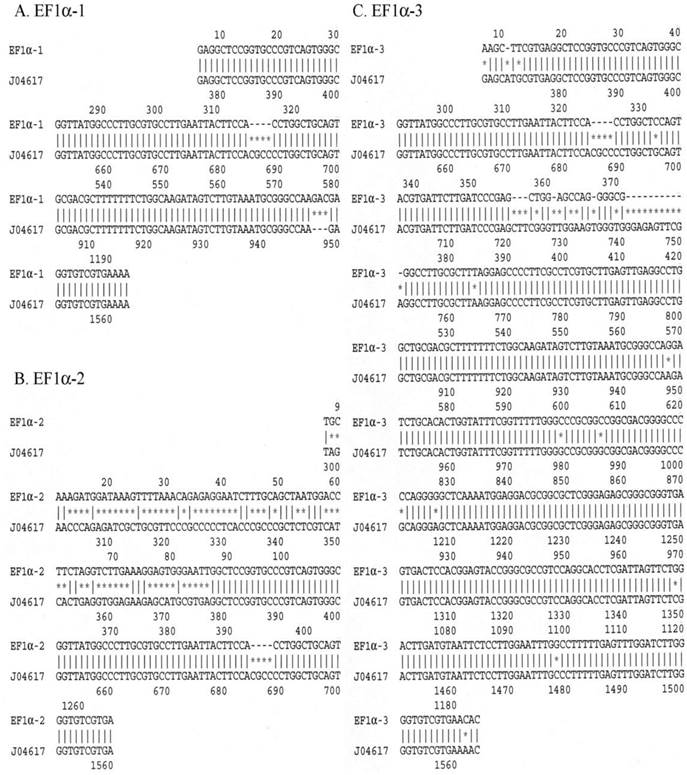
EF1α promoter sequence analysis
To better appreciate possible reasons for the significantly different expression seen from the EF1α-3 promoter, we sequenced all three EF1α promoters in their entirety. We also searched a human EF1α gene in GenBank (NCBI website, accession number: J04617)(17) and used this sequence to compare with the sequences obtained from the EF1α-1, EF1α-2 and EF1α-3 promoters studied here. Differences between the J04617 sequence and the EF1α-1, EF1α-2 or EF1α-3 promoters are shown in Figure 3. The EF1α-1 promoter (panel A) started at nt 374 in J04617. There were two differences between these sequences. One was from nt 685 to 688 in the J04617, i.e. these four base pairs (CGCC) were missing in EF1α-1. The other was located between nt 948 and 949 in J04617; EF1α-1 had three extra base pairs, GAC, inserted. Panel B shows that the EF1α-2 promoter exhibited two differences from the J04617 sequence in GenBank. First, in EF1α-2 there were 81 extra base pairs upstream of J04617 promoter sequence. After searching in GenBank, these 81 base pairs were identified as being derived from the SV40 small antigen. Second, as in EF1α-1, four base pairs (CGCC) were absent in EF1α-2 from nt 685 to 688 in J04617. Panel C shows the comparison between J04617 and the EF1α-3 promoter, and shows three types of differences between them. First, there were thirteen point mutations found in the EF1α-3 promoter, spread throughout the sequence. Second, the same four base pairs (CGCC; nt 685-688) missing in EF1α-1 and EF1α-2 also were absent in the EF1α-3 promoter. Third, the sequence in EF1α-3 following nt 357 showed multiple differences from the comparable region of J04617 (from nt 721 to 751). Is there any transcription factor binding site in these 30 base pairs? Two programs were used to search the binding sites. Seventeen potential transcription factor binding sites, p300, Egr-3, EIIαE-A, R2, Elk-1, VDR, WT1 I, STAT5A, c-Myb, HMG I(Y), STAT4, c-Ets-1, NFI/CIF, Ik-1, MAZ, HSF1 (long) and HSF1 (short) were found by PROMO (18, 19). Using TFBIND (20) program, we found 23 potential transcription factor binding sites, SP1, c-Myb, Elk1, RREB1, p300, AP2, NGFIC, EGR1, EGR2, EGR3, MZF1, ZID, LYF1, AHRARNT, AP4, NFKB, NFKAPPAB50, HSF1, HSF2, ARP1, E2F, STAF and PAX5. These indicate that these 30 base pairs could play important roles in the regulation of EF1α promoter activity.
Comparison of the sequences from the EF1α-1, EF1α-2 and EF1α-3 promoters with that of J04617 (the EF1α sequence from GenBank; accession number in NCBI). A. EF1α-1 promoter; B. EF1α-2 promoter; C. EF1α-3 promoter. Each “*” indicates a sequence difference from that of J04617.
Thus, the sequences of the EF1α-1 and EF1α-2 promoters are quite similar, except for the 81 bp SV40 small antigen sequence at the 5′-end of EF1α-2. However, this extra sequence apparently had little effect on driving transgene expression. Conversely, the sequence differences observed in the EF1α-3 promoter, particularly the first and third ones described above, likely accounted for the considerably lower activity observed with this promoter. The EF1α promoter should have broad activity in multiple cell types and therefore might be widely useful for applications in human gene transfer (6). All three of the promoters studied here are called EF1α promoters in the literature, but they are not equivalent. Clearly, the use of the EF1α-3 promoter by investigators could result in dramatically lower transgene expression.
Acknowledgements
Our research is supported by the Division of Intramural Research, National Institute of Dental and Craniofacial Research, NIH.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Curr Opin Biotechnol. 1999;10:440-447
2. Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439-451
3. Linthout SV, Collen D, Geest BD. Effect of promoters and enhancers on expression, transgene DNA persistence, and hepatotoxicity after adenoviral gene transfer of human apolipoprotein A-I. Hum Gene Ther. 2002;13:829-840
4. Gambotto A, Dworacki G, Cicinnati V. et al. Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: identification of an H2-Kd-restricted CTL epitope. Gene Ther. 2000;7:2036
5. Oh JH, Sohn HY, Kim JM. et al. Construction of multi-purpose vectors, pCNS and pCNS-D2, are suitable for collection and functional study of large-scale cDNAs. Plasmid. 2004;51:217
6. Zheng C, Baum BJ. Evaluation of viral and mammalian promoters for use in gene delivery to salivary glands. Mol Ther. 2005;12:528-536
7. Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322
8. Brown AM, Rusnock EJ, Sciubba JJ, Baum BJ. Establishment and characterization of an epithelial cell line from the rat submandibular gland. J Oral Pathol Med. 1989;18:206-213
9. Yanagawa T, Hayashi Y, Nagamine S. et al. Generation of cells with phenotypes of both intercalated duct-type and myoethelial cells in human parotid gland adenocarcinoma clonal cells grown in athymic nude mice. Virchows Arch B Cell Pathol Incl Mol Pathol. 1986;51:187-195
10. Shirasuna K, Sato M, Miyasaki T. A neoplastic epithelial duct cell line established from an irradiated human salivary gland. Cancer. 1981;48:745-752
11. Graham FL, Smiley J, Russel WC, Nairn R. Characteristics of a human cell lines transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59-74
12. Baker A, Saltik M, Lehrmann H. et al. Polyethylenimine (PEI) is a simple, inexpensive and effective reagent for condensing and linking plasmid DNA to adenovirus for gene delivery. Gene Ther. 1997;4:773-782
13. Meunier-Durmort C, Grimal H, Sachs LM. et al. Adenovirus enhancement of polyethylenimine-mediated transfer of regulated genes in differentiated cells. Gene Ther. 1997;4:808-814
14. Mastrangeli A, O'Connell B, Aladib W. et al. Direct in vivo adenovirus-mediated gene transfer to salivary glands. Am J Physiol. 1994;266(G):1146-1155
15. Delporte C, O'Connell BC, He X. et al. Increased fluid secretion after adenoviral-mediated transfer of the aquaporin-1 cDNA to irradiated rat salivary glands. Proc. Natl. Acad. Sci. U S A. 1997;94:3268-3273
16. He X, Goldsmith CM, Marmary Y. et al. Systemic action of human growth hormone following adenovirus-mediated gene transfer to rat submandibular glands. Gene Ther. 1998;5:537-541
17. Uetsuki T, Naito A, Nagata S, Kaziro Y. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1 alpha. J Biol Chem. 1989;264:5791-5798
18. Messeguer X, Escudero R, Farre D. et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333-334
19. Farre D, Roset R, Huerta M. et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651-3653
20. Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622-630
Author contact
![]() Corresponding author: Dr. Changyu Zheng, MPTB/NIDCR/NIH, Bldg. 10, Room 1A01, MSC-1190, 9000 Rockville Pike, Bethesda, MD 20892-1190. Tel: (301)594-1924 Fax: (301)402-1228 Email: czhengnih.gov.
Corresponding author: Dr. Changyu Zheng, MPTB/NIDCR/NIH, Bldg. 10, Room 1A01, MSC-1190, 9000 Rockville Pike, Bethesda, MD 20892-1190. Tel: (301)594-1924 Fax: (301)402-1228 Email: czhengnih.gov.

 Global reach, higher impact
Global reach, higher impact