Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(2):215-221. doi:10.7150/ijms.7231 This issue Cite
Research Paper
Gene Expression Analysis of TREM1 and GRK2 in Polymorphonuclear Leukocytes as the Surrogate Biomarkers of Acute Bacterial Infections
Department of Microbiology & Immunology, Teikyo University School of Medicine. Tokyo 173-8605, Japan
Received 2013-7-23; Accepted 2013-12-11; Published 2014-1-10
Abstract
Objective: In the acute stage of infectious diseases such as pneumonia and sepsis, sequelae hypercytokinemia and cytokine storm are often observed simultaneously. During bacterial infections, activated polymorphonuclear leukocytes (PMNs) cause inflammation and organ dysfunction in severely ill patients. Gene expression of the triggering receptor on myeloid cells (TREM)-1 and G-coupled-protein receptor kinase (GRK)-2 in PMNs isolated from patients was analysed to identify genes correlated with the severity of pathophysiological conditions.
Methods: mRNA levels of TREM1 and GRK2 in the PMNs from 26 patients (13 with pneumonia, 5 with severe sepsis, and 8 with septic shock) were analysed by using quantitative real-time PCR. The synthesised soluble form (s)TREM-1 was incubated with normal PMNs to investigate its biological functions in vitro.
Results: Copies of TREM1 transcript were 0.7- to 2.1-fold higher in patients with pneumonia compared to those of normal subjects; the average fold-change was 1.1-fold. The mRNA levels of patients suffering from severe sepsis and septic shock were 0.34- and 0.33-fold lower compared to those of healthy subjects, respectively. TREM1 mRNA levels in 5 of 26 patients in convalescent stages recovered to normal levels. The mRNA levels of GRK2 in the PMNs of patients were also downregulated. The synthesised sTREM-1 upregulated the mRNA levels of TREM1 in normal PMNs.
Conclusions: TREM1 mRNA levels were inversely correlated with the severity of pathophysiological conditions in acute bacterial infections. The gene expression levels of TREM1 in PMNs isolated from patients with bacterial infections may be used as a surrogate biomarker for determining the severity.
Keywords: Pneumonia, Sepsis, Polymorphonuclear leukocytes, real-time PCR, TREM1, GRK2
Introduction
The World Health Organization (WHO) estimates that lower respiratory tract infections account for nearly 35% of all deaths from infectious diseases, causing an annual mortality of nearly 4 million adults and children. Thus, bacterial pneumonia is a significant cause of mortality worldwide [1].
Severe sepsis and septic shock are common among critically ill patients, with a reported incidence ranging from 50 to 300 cases per 100,000 people and mortality rates ranging from 25% to 70% when complicated by shock and multiple organ failure [2-5]. Furthermore, some infected individuals die from secondary infections during the phase of sepsis-associated immunosuppression (SAIS) [6].
In acute bacterial infections such as pneumonia and sepsis, PMNs are critical effector cells of the innate immune system, which protect the host by migrating to inflammatory sites and killing pathogenic microbes. PMN migration is regulated by signaling mechanisms activated by chemokines, which act on chemokine receptors in order to direct PMN migration along a concentration gradient [7].
Desensitization of chemokine receptors within the G protein-coupled receptor (GPCR) family is an important determinant of the intensity and duration of agonist stimulation [8]. Receptor desensitisation regulates not only the number of migrating PMNs, but also their motility and ability to stop upon contact with pathogens or target cells [8]. GPCR kinases (GRKs), specific kinases that interacting with GPCRs, induce receptor phosphorylation and thereby signal GPCR desensitization [9]. The chemokine macrophage inflammatory protein (MIP)-2 induces GRK2 expression in PMNs through the phosphoinositide-3-kinase (PI3K)-γ signaling pathway and lipopolysaccharide (LPS)-activated signaling through the Toll-like receptor (TLR)-4 pathway, which transcriptionally downregulates the expression of GRK-2 in response to MIP-2 [10]. TLR4 modulation of the expression of chemokine receptors on the surface of PMNs following the downregulation of GRK2 expression is a critical determinant of PMN migration.
Triggering receptor expressed on myeloid cells (TREM)-1 is a recently discovered member of the immunoglobulin (Ig) superfamily [11]. TREM-1 is an amplifier of the immune response, which strongly potentiates the activation of PMNs in response to microbial products [12]. Soluble TREM-1 (sTREM-1) levels in patients' plasma are superior indicators of sepsis compared to C-reactive protein (CRP) and procalcitonin (PCT) levels [13-16]. The function of TREM-1 and sTREM-1 is to modulate the inflammatory response during pneumonia and sepsis [17].
PMNs in peripheral blood constitute an accessible source of clinically relevant information, and gene expression profiling of these cells is an emerging strategy for diagnosing and monitoring infections. Elucidating the gene expression patterns of TREM1 and GRK2 in the PMNs of patients with bacterial infections has led to a better understanding of the underlying mechanisms of infectious diseases and responses to treatment.
Methods
Patients and controls
Thirteen patients (12 males and 1 female) with pneumonia were followed-up using the criteria of The Japanese Respiratory Society [18]. The median age of patients with moderate cases was 60 years, and the median age of those with severe illness was 71 (Table 1). Thirteen patients (3 males and 10 females) with sepsis were followed-up by the definitions [19], and the median ages of patients with severe sepsis and septic shock were 72 years and 70 years, respectively (Table 1). All patients were hospitalised at the Teikyo University Hospital. The causative organisms were isolated from the sputum of pneumonia patients or whole blood of sepsis patients. All organisms were cultured and identified in the Clinical Laboratory Department of the hospital. The protocol was approved by the Ethical Review Committee at the Teikyo University School of Medicine (No. 07-104), and all participants gave written informed consent.
Clinical characteristics of patients with acute bacterial infections.
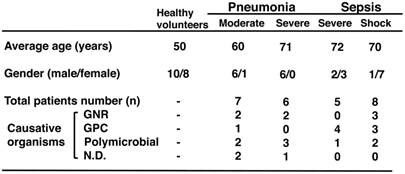
GNR, Gram-negative rod; GPC, Gram-positive coccal; and ND, not detected. The total number of patients was 23.
PMN preparation
PMNs from patients and healthy volunteers were isolated from the peripheral blood [20]. Briefly, whole blood (20 mL) was mixed with 4.5% dextran solution, and the mixture was allowed to stand at room temperature for 40 minutes. The leukocyte-rich plasma was then centrifuged at 400g on a Ficoll-Paque Plus gradient (Amersham Bioscience, WI, USA) for 20 minutes. To lyse erythrocytes, a hypotonic (0.2%) saline solution was used. The osmolality of the mixture was then restored using a hypertonic (1.6%) saline solution. The PMNs were adjusted to a final concentration of 1 x 107 cells/mL in Hank's balanced salt solution (HBSS). All volunteers were healthy adults (10 males and 8 females) with ages ranging from 25 years to 58 years (mean age, 50 years).
RNA isolation
Total RNA was extracted from the PMNs by using the RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The quantity and quality of total RNA samples were determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany).
Complementary DNA synthesis
Total RNA was reverse transcribed to cDNA by using SuperScript VILO cDNA synthesis kit (Invitrogen Life Technologies, CA, USA). Briefly, 1 μg of total RNA was incubated with 2.5 μM oligo (dT)20, and 50 ng random hexamers, and 200 U SuperScript III RT enzyme in a 20-μL reaction volume at 42ºC for 60 minutes followed by 50ºC for 20 minutes. The reactions were terminated by heating at 85ºC for 5 minutes.
Quantitative real-time PCR (qPCR) analysis
Gene expression levels of TREM1 (GenBank accession no. MN_018643.2) and GRK2 (GenBank accession no. MN_001619.3) in PMNs were quantified using the StepOne real-time PCR system (Applied Biosystems, CA, U.S.A.). Complementary DNA was amplified with SYBR Green by using the Power SYBR Green PCR master mix (Invitrogen, CA, USA). Briefly, qPCR was performed for TREM1 and the housekeeping gene, ACTB (GenBank accession no. MN_001101.1). PCR primer sets were designed using the Primer3 web site (http://bioinfo.ut.ee/primer3/), and the primer sequences are shown in Table 2 [21]. The following cDNA amplification program was used: 95ºC for 10 minutes; and 40 cycles of 95ºC for 15 s, 60ºC for 1 minute. All PCR reactions were performed in 20-μL reaction volumes comprising the following components: 5 μL cDNA solution, 0.9 U Ampli Taq Gold DNA polymerase, 1 x reaction buffer (20 mM Tris-HCl pH 8.4, 3 mM MgCl2, 200 μM dVTPs [mixture of dATP, dCTP, and dGTP], 400 μM dUTP, 500 nM ROX reference dye, and 0.6 U uralic glycosylase), and 200 nM primers. TREM1 mRNA expression levels in PMNs were normalised to the gene expression levels of ACTB. Fold changes of PMN TREM1 mRNA levels between patients and controls were determined using the Sequence Detection System (SDS) software (Applied Biosystems).
Quantitative real-time PCR primer sets.

F, forward primer; and R, reverse primer.
Oligopeptide synthesis of sTREM-1
A segment of sTREM-1 (length 50 amino acid residues: #18-67) was synthesised by OPERON (Tokyo, Japan). The sequence was designed using a 3D software and is shown below: NH2-LRAATKLTEEKYELKEGKTLDVKCDYTLEKFASSQKAWQIIRDGEMPKTL-COOH. The synthesised sTREM-1 was incubated with normal PMNs at 37ºC for 1 hour. The reaction mixture contained 0.2% (v/v) human serum (final concentration).
Statistical analysis
All P values were determined using nonparametric unpaired or paired t-tests (two-tailed) in Excel 2011 (Microsoft Corporation, Tokyo, Japan). We considered P < .05 to be significant, and the degree of significance was indicated as follows: **, P < .01.
Results
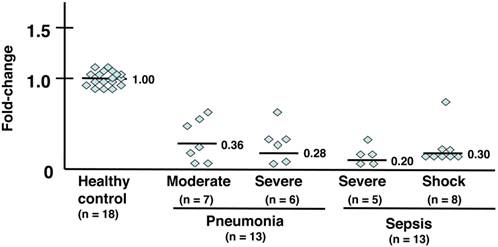
Expression analysis of the G-protein-coupled receptor kinase GRK2 (Fig. 1 and Table 3a)
The gene expression levels of GRK2 in PMNs were downregulated in all patients with acute bacterial infections such as pneumonia and sepsis (Table 3a). The following average fold-changes were observed in patients (Fig. 1): moderate pneumonia, 0.36; severe pneumonia, 0.28; severe sepsis, 0.20; and septic shock, 0.30.
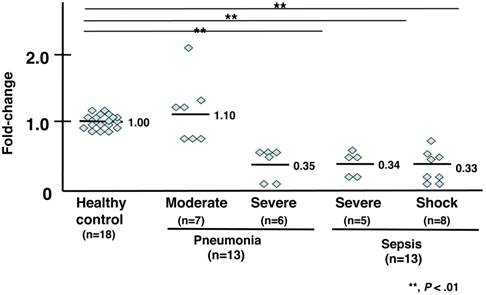
Expression analysis of the triggering receptor TREM1 (Fig. 2 and Table 3a)
The gene expression levels of TREM1 in 4 of 7 patients with moderate pneumonia were higher (an average fold-change of 1.10) than those in healthy subjects. Six patients with severe pneumonia showed 0.50-fold lower expression levels of TREM1 compared to that of healthy volunteers, and the levels in 2 patients with severe pneumonia were 0.10-fold lower than those of healthy volunteers. The average fold-change was 0.35. The mRNA levels of TREM1 were lower in 5 patients with severe sepsis compared to those of control subjects. The average fold-change was 0.34. The mRNA levels of TREM1 of 8 patients in septic shock were 0.33-fold lower than those of healthy volunteers.
TREM1 mRNA levels of patients between acute and convalescent stages of bacterial infectious (Table 3b)
The TREM1 mRNA levels of 5 of 26 patients between acute and convalescent stages were examined. TREM1 mRNA levels in 1 moderate pneumonia patient (patient I.D.# 10 and 10C*), 1 severe pneumonia patient (patient I.D.# 16 and 16C*), and 3 septic shock patients (patient I.D.# 22 and 22C*, 23 and 23C*, 24 and 24C*) recovered to those of healthy subjects.
mRNA expression analysis of GRK2. Individual values were plotted, and each bar represents the mean of the values. Eighteen healthy subjects served as a control, with an average mRNA expression level of 1.0. Seven patients with moderate pneumonia had a 0.36-fold average change in mRNA expression levels. Six patients with severe pneumonia had a 0.28-fold average change in mRNA levels. Five patients with severe sepsis had a 0.20-fold average change in mRNA levels. Eight patients in septic shock had a 0.30-fold average change in mRNA levels. The statistical significance was determined using the Mann-Whitney U test. **, P < .01 compared with healthy subjects. Data are representative of at least 3 separate experiments.
Fold-changes of mRNA expression levels of TREM1 and GRK2 in infected patients.
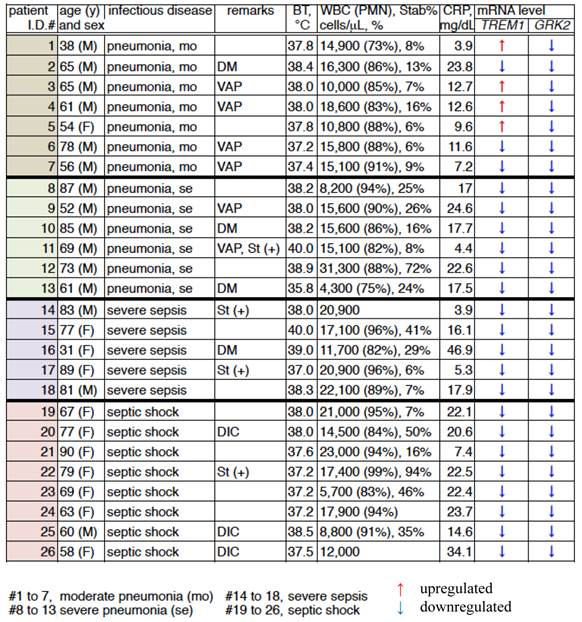
BT, body temperature; WBC, white blood cells; CRP, C-reactive protein; DM, diabetes mellitus; VAP, ventilator-associated pneumonia; St(+), steroid treatment; and DIC, disseminated intravascular coagulation.
mRNA expression analysis of TREM1. Individual values were plotted, and each bar represents the mean of the values. Eighteen healthy subjects served as a control, with an average mRNA expression level of 1.0. Seven patients with moderate pneumonia had a 1.10-fold average change in mRNA levels. Six patients with severe pneumonia had a 0.35-fold average change in mRNA levels. Five patients with severe sepsis had a 0.34-fold average change in mRNA levels. Eight patients in septic shock had a 0.33-fold average change in mRNA levels. The statistical significance was determined using the Mann-Whitney U test. **, P < .01 compared to healthy subjects. Data are representative of at least 3 separate experiments.
Comparison of the TREM1 mRNA levels in the PMNs of patients in between the acute and convalescent stages of bacterial infection.
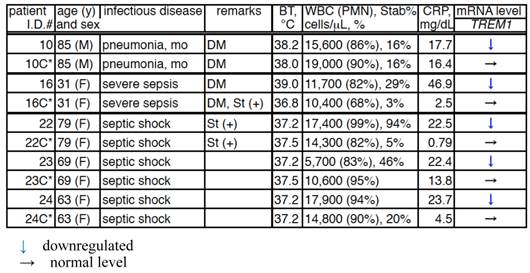
C*, convalescent; BT, body temperature; WBC, white blood cells; CRP, C-reactive protein; DM, diabetes mellitus; and St(+), steroid treatment.
TREM1 mRNA levels in normal PMNs treated with synthesised sTREM-1 with human serum (Fig. 3)
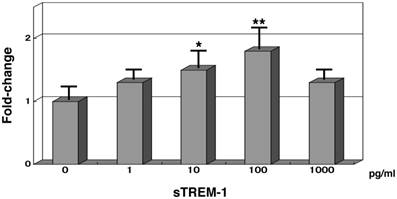
Synthesised sTREM-1 oligopeptide (50 residues) was administered to normal PMNs and then incubated for 1 hour with 0.2% human serum. The TREM1 mRNA expression levels were subsequently measured. When 1 pg/mL of sTREM-1 was added to normal PMNs, the TREM1 mRNA levels were 1.3-fold higher than normal expression levels. TREM1 mRNA expression levels increased to 1.8-fold higher than normal expression levels in the presence of 100 pg/mL of sTREM-1 oligopeptide. However TREM1 mRNA expression levels was 1.3-fold higher than normal expression levels in the presence of 1,000 pg/mL of sTREM-1 oligopeptide.
Gene expression analysis of TREM1 in normal PMNs with synthesised soluble TREM-1 (sTREM-1) in the presence of human serum. mRNA expression levels of 0 pg/mL sTREM-1 were 1.0. Each samples contained 0.2% human serum. *, P < .05; and **, P < .01 compared with healthy subjects. Error bars represent the SEM. Data are representative of at least 3 separate experiments.
Discussion
Among phagocytes, PMNs are remarkable for their migratory capacity. Upon activation by chemoattractants, chemokines, or inflammatory products, PMNs rapidly migrate to sites of infection [22]. Pathogen-associated molecular patterns (PAMPs) are recognised as molecular signatures by pattern recognition receptors (PRRs) that are predominantly expressed on PMN surface [23].
The findings of GRK2 gene expression analysis did not correlate with the body temperature (BT), white blood cell (WBC) count, C-reactive protein (CRP) level, age, and gender of patients. The mRNA levels of GRK2 were downregulated in the PMNs of all patients with bacterial infections, indicating that PMN migration was suppressed. Indeed, the ability of PMN migration of our patients with severe infections was inhibited (data not shown). In our previous gene expression analysis of PMNs from patients with acute bacterial infections, 7 immunomodulated genes (i.e., TLR2, TLR4, CD14, TNFA, IL6, IL8Rs, and MAC-1) did not correlate with the conditions of 23 infected patients [21].
However, the inverse correlation of TREM1 mRNA levels with the severity of pathophysiological conditions of acute bacterial infections was overlooked. Protein expression of TREM-1 is also upregulated in phagocytic cells in the presence of pathogens, and sTREM-1 is released into circulating blood during infection [24].
In this study, the average expression levels of TREM1 in moderate and severe pneumonia patients were 1.10- and 0.35-fold, respectively. The mRNA levels of TREM1 in patients suffering from severe sepsis and septic shock were 0.34- and 0.33-fold, respectively (Fig. 2 and Table 3a). In 5 of 26 cases, in which patients were in the convalescent stage of acute bacterial infections, TREM1 mRNA expression levels recovered to normal levels (Table 3b). Therefore, monitoring the mRNA levels of TREM1 in the PMNs of patients could possibly reflect the effect of treatment.
The serum levels of CRP and PCT are used as markers for inflammatory and infectious diseases worldwide. These data are easy and convenient to measure because of the quantity of protein available in the serum of patients during routine work [25]. However, the mRNA levels of TREM1 in patients with acute bacterial infections are more specific for their pathophysiological conditions and even require total RNA extraction and cDNA synthesis. Recently, these steps have become quick and easy with the use of automatic equipment [26]. The mRNA expression levels of TREM1 in circulating PMNs may reflect the severity of a bacterial infection.
Moreover, we also investigated the effects of synthesised sTREM-1 on TREM1 gene expression in normal PMNs. The mRNA levels of TREM1 were upregulated using 50 amino-acid residues of sTREM-1 in a dose-dependent manner (range: 0-100 pg/mL). However the gene expression levels of TREM1 was 1.3-fold higher than normal expression levels in the presence of 1,000 pg/mL of sTREM-1 oligopeptide. In the acute stage of bacterial infections, >100 pg/mL of sTREM-1 releases from PMNs and monocytes into the serum of patients [13]. During the conditions, such as severe sepsis and septic shock, gene expression levels of TREM1 in patients' PMNs were downregulated (Table 3a and Fig. 2).
The full-length of sTREM-1 has been reported to amplify inflammation in animals [27]. Soluble TREM-1 may be a key factor that exacerbates acute infectious disorders because blocking TREM-1 signaling with a sTREM-1-IgG fusion protein has reduced hyperinflammatory responses and death in animal models [28].
The relevance of our findings is limited to PMNs, and the interactions between the alteration of TREM1 gene expression and protein synthesis have not been fully elucidated. Furthermore, it is unclear whether similar changes occur in other leukocyte subtypes (eg, lymphocytes, monocytes, M1 and M2 macrophages). Future studies on these cell types are required if we are to gain a more complete understanding of the host response to bacterial infections.
Acknowledgements
We thank our patients for agreeing to participate in this study. We are also grateful to Ms. C. Miyazaki for her technical assistance.
Financial support for this study was provided by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21591300).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mizgerd JP. Lung infection: a public health priority. PLoS Med. 2006;3:e76
2. Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Cir Care Med. 2001;29:1303-10
3. Russel JA. Management of sepsis. N Engl J Med. 2006;355:699-713
4. Dombrovskiy VY, Martin AA, Sunderram J, Paz HL. Rapid increase in hospitalization and mortality rates for severe sepsis in the United States: a trend analysis from 1993 to 2003. Crit Care Med. 2007;35:1244-50
5. Dellinger RP, Levy MM, Carlet JM, Bion J, Parker MM, Jaeschke R. et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296-327
6. Hoetzenecker W, Echtenacher B, Guenova E, Hoetzenecker K, Woelbing F, Bruck J. et al. ROS-induced ATF3 causes susceptibility to secondary infections during sepsis-assiciated immunosuppression. Nat Med. 2012;18:128-34
7. Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565-568
8. Lefkowitz RJ. G protein-coupled receptors. III. New roles for receptor kinases and β-arrestins in receptor signaling and desensitization. J Biol Chem. 1998;273:18677-80
9. Gainetdinov RR, Bohn LM, Walker JK, Laporte SA, Macrae AD, Caron MG. et al. Muscarinic supersensitivity and impaired receptor desensitization in G protein-coupled receptor kinase 5-deficient mice. Neuron. 1999;24:1029-36
10. Fan J, Malik AB. Toll-like receptor-4 (TLR4) signaling augments chemokine-induced neutrophil migration by modulating cell surface expression of chemokine receptors. Nature Med. 2003;9:315-21
11. Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3:445-53
12. Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164:4991-5
13. Gibot S, Cravoisy A, Kolopp-Sarda MN, Bene MC, Faure G, Bollaert PE. et al. Time-course of sTREM (soluble triggering receptor expressed on myeloid cells)-1, procalcitonin, and C-reactive protein plasma concentrations during sepsis. Crit Care Med. 2005;33:792-6
14. Gibot S, Cravoisy A, Levy B, Bene MC, Faure G, Bollaert PE. Soluble triggering receptor expressed on myeloid cells and the diagnosis of pneumonia. N Engl J Med. 2004;350:451-8
15. Gibot S, Kolopp-Sarda MN, Bene MC, Cravoisy A, Levy B, Faure GC. et al. Plasma level of a triggering receptor expressed on myeloid cells-1: its diagnostic accuracy in patients with suspected sepsis. Ann Intern Med. 2004;141:9-15
16. Gibot S, Massin F, Le Renard P, Bene MC, Faure GC, Bollaert PE. et al. Surface and soluble triggering receptor expressed on myeloid cells-1: expression patterns in murine sepsis. Crit Care Med. 2005;33:1787-93
17. Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat Immunol. 2006;7:1266-73
18. Kohno S. The JRS Guidelines for the management of community-acquired pneumonia in adults. Kohno S. (Ed.), The Japanese Respiratory Society Publishing, Tokyo, Japan. 2008
19. Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA. et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644-55
20. Ubagai T, Tansho S, Ito T, Ono Y. Influences of aflatoxin B1 on reactive oxygen species generation and chemotaxis of human polymorphonuclear leukocytes. Toxicol in Vitro. 2008;22:1115-20
21. Ubagai T, Tansho S, Ieki R, Ono Y. Evaluation of TREM1 gene expression in circulating Polymorphonuclear leukocytes and its inverse correlation with the severity of pathophysiological conditions in patients with acute bacterial infections. Jpn J Infect Dis. 2012;65:376-82
22. Nathan C. Neutrophils and immunity: Challenges and opportunities. Nat Rev Immunol. 2006;6:173-82
23. Kawai T, Akira S. The roles of TLRs, RLRs, and NLRs in pathogen recognition. Int Immumol. 2009;21:317-37
24. Gibot S, Cravoisy A. Soluble form of the triggering receptor expressed on myeloid cells-1 as a marker of microbial infection. Clin Med & Res. 2004;2:181-7
25. Mitsuma SF, Mansour MK, Dekker JP, Kim J, Rahman Z, Tweed-Kent A. et al. Promising new assay and technologies for the diagnosis and management of infectious diseases. Clin Infect Dis. 2013;56:996-1002
26. Reinhart K, Bauer M, Riedemann NC, Hartog CS. New approaches to sepsis: molecular diagnostics and biomarkers. Clin Microbiol Rev. 2012;25:609-34
27. Colonna M, Facchetti F. TREM-1 (triggering receptor expressed on myeloid cells): a new player in acute inflammatory responses. J Infect Dis. 2003;187(Suppl. 2):S397-S401
28. Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103-7
Author contact
![]() Corresponding author: Tsuneyuki Ubagai. Department of Microbiology & Immunology, Teikyo University School of Medicine, Kaga 2-11-1, Itabashi-ku, Tokyo 173-8605, Japan. Phone: +81-3-3964-1211; Fax: +81-3-5375-5284; E-mail: ubatteikyo-u.ac.jp
Corresponding author: Tsuneyuki Ubagai. Department of Microbiology & Immunology, Teikyo University School of Medicine, Kaga 2-11-1, Itabashi-ku, Tokyo 173-8605, Japan. Phone: +81-3-3964-1211; Fax: +81-3-5375-5284; E-mail: ubatteikyo-u.ac.jp

 Global reach, higher impact
Global reach, higher impact