Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(2):164-171. doi:10.7150/ijms.6745 This issue Cite
Research Paper
Aberrant DNA Methylation of G-protein-coupled Bile Acid Receptor Gpbar1 (TGR5) is a Potential Biomarker for Hepatitis B Virus Associated Hepatocellular Carcinoma
1. Department of Hepatology, Qilu Hospital of Shandong University, Jinan 250012, China
2. Institute of Hepatology, Shandong University, Jinan 250012, China
3. Department of Ultrasound, the General Hospital Jinan Military Region, Jinan 250031, China
Received 2013-5-23; Accepted 2013-12-24; Published 2014-1-7
Abstract
Background: G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a newly identified liver tumor suppressor in carcinogenesis. This present study was therefore to determine the potential value of serum TGR5 promoter methylation in identifying hepatocellular carcinoma (HCC) from chronic hepatitis B (CHB) patients.
Methods: The circulating cell-free DNA (cfDNA) was extracted from a retrospective dataset including 160 HCC, 88 CHB and 45 healthy controls (HCs). Methylation status of TGR5 promoter was examined by methylation-specific polymerase chain reaction (MSP).
Results: Hypermethylation of the TGR5 promoter occurred significantly more frequent in HCC (77/160, 48.13%) than CHB (12/88, 13.64%; p<0.01) and HCs (2/45, 4.44%; p<0.01). The methylation rate of TGR5 in HCC patients ≥60 years old was significantly higher than those <60 years old (p<0.05). Alpha fetoprotein (AFP) had sensitivity of 58.13%, 30.63% and 24.38% at cut-off points of 20, 200 and 400ng/ml respectively; while TGR5 methylation combined AFP had sensitivity of 81.25%, 68.13% and 65%. AFP had specificity of 47.73%, 92.05% and 98.86% at cut-off points of 20, 200 and 400ng/ml respectively; while TGR5 methylation combined AFP had specificity of 38.64%, 78.41% and 85.23%. AFP had Youden index of 0.06, 0.23 and 0.23 at cut-off points of 20, 200 and 400ng/ml respectively; while TGR5 methylation combined AFP had Youden index of 0.20, 0.47 and 0.50.
Conclusions: Our findings strongly suggested the combination of serum TGR5 promoter methylation and AFP enhanced the diagnostic value of AFP alone in discriminating HCC from CHB patients.
Keywords: TGR5, DNA methylation, Hepatocellular carcinoma, MSP, serum
Introduction
Liver cancer is the fifth most common cancer for men and the seventh for women worldwide [1]. Hepatocellular carcinoma (HCC) represents the major histological subtype and accounts for 70% to 85% of liver cancers worldwide [2]. Accumulating evidences showed that the incidence of HCC was still rising and 5-years survival of HCC was as low as about 0%-10% [3]. Until now, the screening and surveillance for HCC mainly depend on serum level of alpha fetoprotein (AFP) and ultrasonography. However, several studies showed that the determination of serum AFP level lack adequate sensitivity and specificity for effective diagnosis and surveillance of HCC [4, 5]. The performance of ultrasonography was influenced by many factors including experience of the examiner, the technology used, the body habitus, the presence of cirrhosis and the size of the tumor [6]. Therefore, new biomarkers for the surveillance and early detection of HCC are urgently needed.
DNA methylation is one of the most common epigenetic mechanisms, which donates the addition of a methyl group to DNA to regulate the expressions of its downstream targeted genes. It is widely spread in human genome and usually occurs at cytosine adjacent to guanine (CpG dinucleotides) [7]. Aberrant DNA methylation of several cancer related genes occurred in most of human malignancy [8, 9]. DNA methylation can be detected in free-floating DNA from dead cancer cells in bodily fluids and can provide novel potential biomarkers for the early diagnosis of human cancers including HCC [10]. Our previous study demonstrated that TFPI2 methylation in serum might be used as a predictor of HCC progression [11]. In recent years, several genes including GSTP1, RASSF1A, APC, and P16 were also found to be aberrantly methylated in serum or tissue of HCC patients and may potentially be used as biomarkers [12-14].
G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a membrane-bound receptor for bile acids and is well known for its role in regulating energy homeostasis, bile acid homeostasis, as well as glucose metabolism [15-17]. TGR5 has been reported to regulate hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) in Mice [18]. A recent study demonstrated that TGR5 activation greatly inhibits proliferation and migration of human liver cancer cells in vitro and the deficiency of TGR5 enhances chemical-induced liver carcinogenesis [19]. These findings identified that TGR5 is a novel tumor suppressor gene and TGR5 might be an attractive therapeutic tool for liver cancer.
The human TGR5 promoter has not been definitely characterized up to date. In our present study, we recognize the region within 2000bp upstream of transcriptional start site as presumptive promoter region for TGR5 gene like several other studies [20, 21]. GeneBank indicates that 37 CpG sites located on TGR5 promoter sequence (Supplementary figure 1). Therefore, methylation of TGR5 gene promoter may occur in tumorigenesis and may be a potential biomarker for the diagnosis of tumor diseases in clinic. In our present study, we investigated promoter methylation of TGR5 gene in the serum of hepatitis B virus (HBV) infected patients with or without HCC and healthy blood donors. Then we evaluated aberrant promoter methylation of TGR5 gene in serum cfDNA as a biomarker for screening HCC from CHB.
Patients and Methods
Patients
160 HBV-infected patients with HCC, 88 CHB patients without HCC and 45 HCs were recruited from April 2012 to April 2013 in the Department of Hepatology, Qilu Hospital of Shandong University. HCC patients were diagnosed according to the 2010 update of the American Association for the Study of Liver Diseases (AASLD) Practice Guidelines for Management of hepatocellular carcinoma [22]. Chronic HBV infection was defined as a positive hepatitis B surface antigen (HBsAg) for at least 6 months prior to the beginning of this study [23]. Within all the 88 CHB patients, 33 were accompanied by cirrhosis. Exclusion criteria included co-infection with human immunodeficiency virus (HIV) or hepatitis C virus (HCV), alcoholic liver diseases, autoimmune liver diseases, non-alcoholic fatty liver diseases (NAFLD) and other causes of chronic liver diseases. All patients gave written informed consents under protocols approved by the local Research and Ethics Committee at Qilu Hospital of Shandong University, in accordance with the guidelines of the 1975 Declaration of Helsinki.
Clinic pathological data collection
Blood samples were taken from each participant who was fasting for more than 12 hours before blood collecting. HBeAg was measured by an automatic analyzer (cobas 6000 analyzer series, Roche Diagnostics, Switzerland). The serum biochemical markers (COBAS integra 800, Roche Diagnostics, Germany) included alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL) and albumin (ALB). Hemostasis markers (ACL TOP 700, Instrument laboratory, USA) included prothrombin time-international normalized ratio (PT-INR). AFP was also measured by an automatic analyzer (COBAS e 601, Roche Diagnostics, Germany). These markers were measured using standard methodologies in Department of Laboratory Medicine, Qilu Hospital, Shandong University.
All of the HCC patients received enhanced CT scan (Discovery CT750, GE, USA) in Department of Medical Imaging, Qilu Hospital, Shandong University. The CT films were examined by one radiologist in Department of Medical Imaging, Qilu Hospital who was unaware of patients' characteristics. The diagnosis of HCC was confirmed histologically in all cases. The HCC tissue specimens were examined by one pathologist in Department of Pathology, Qilu Hospital who were unaware of patients' characteristics.
Serum DNA Extraction and Sodium Bisulfite Modification
A minimum volume of 400 μL serum was obtained from every participant. Serum DNA was extracted by the QIAamp DNA Blood Mini Kit (Qiagen, Germany) according to the ''Protocol: DNA Purification from Blood or Body Fluids'' recommended by the manufacturer. DNA concentration was then determined by the Eppendorf Biophotometer (Brinkmann Instruments, Westbury, NY, USA). The extracted DNA was eluted in a total volume of 200μL sterile water and stored at -20 °C until sodium bisulfate modification.
DNA bisulfate modification was performed by EZ DNA Methylation-Gold KitTM (Zymo Research, USA) according to the manufacturer's instructions. A final volume of 20 μL modified DNA was obtained, and was either used immediately as a template for MSP or stored at -20 °C.
Methylation-Specific Polymerase Chain Reaction (MSP)
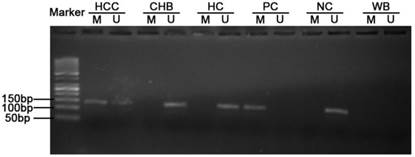
Methylated and unmethylated primers specific for TGR5 promoters were designed using MethPrimer according to the recommended criteria [24]. The reasons for choosing this area included: (i) the primers contained at least one CpG site at the most 3'-end; (ii) the primers in M pair and U pair contained the same CpG sites within their sequence; (iii) two sets of primers had similar product Tm values, which is 66.3oC for M pair and 66.0oC for U pair. Then the selected primer sets were used to amplify the bisulfite modified DNA in our study (Table 1). The M pair primers amplified -1958 to -1821 site of the 5'-UTR of the TGR5 gene (+1 for the transcriptional start site). Meanwhile, the U pair primers amplified -1955 to -1820 site of the 5'-UTR of the TGR5 gene (Supplementary figure 1). MSP was performed in a total volume of 25μL containing 1μL bisulfite-treated DNA, 0.5μL of each primer (10 μM), 10.5μL nuclease-free water, and 12.5μL Premix Taq (Zymo Research, USA), which consisted of Taq DNA polymerase, reaction buffer, and deoxynucleotide triphosphate mixture. The PCR protocol was composed of an initial denaturation at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 30s, annealing at 56 °C for 40 s, and primer extension at 72 °C for 40 s; PCR cycles were followed by final extension at 72 °C for 10 min. Human DNA, treated in vitro with SssI methylase and bisulfate converted (Qiagen, Germany), was used as positive control. DNA from normal lymphocytes was used as negative control. Water bland without DNA was used as in each round of PCR. PCR products were then electrophoresed on a 2 % agarose gel, stained with ethidium bromide, and visualized under UV illumination. Each MSP was repeated for three times.
Statistical analysis
Quantitative variables were expressed as the median (centile 25; centile 75). Categorical variables were expressed as number (percentage). Statistical analyses of the data were performed with SPSS version 16.0 (Chicago, IL, USA). χ2 test or Fisher's exact test were used to compare the categorical variables. Spearman correlation coefficients were calculated to evaluate correlations between serum AFP and TGR5 methylation. A 2-tailed p value less than 0.05 was considered statistically significant.
Primers for MSP of the TGR5 gene
| Primer name | Primer sequence (5'-3') | Product size (bp) | Annealing temp (°C) |
|---|---|---|---|
| M | F: TTTTTGTTTAAATGGTTTTTATTGTC | 138 | 56 |
| R: CACTTCCTATTAAAATCTTAACGTC | |||
| U | F: TTGTTTAAATGGTTTTTATTGTTGA | 136 | 56 |
| R: CCACTTCCTATTAAAATCTTAACATC |
M methylated sequence, U unmethylated sequence, F forward, R reverse
Results
General characteristics
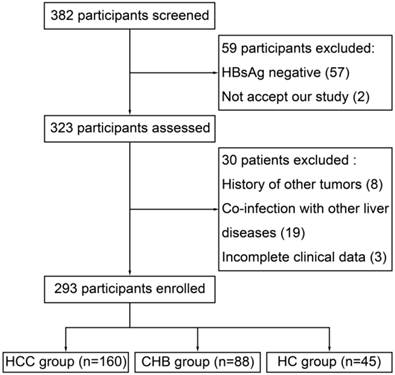
From April 2012 to April 2013, 382 participants were screened in Department of Hepatology, Qilu Hospital. 59 participants were excluded because of HBsAg negative (57) and not accept our study (2). Then, 8 patients were excluded for history of other tumor, 19 for co-infection with other liver diseases and 3 for incomplete clinical data, which left 293 participants enrolled (Figure 1). Within the enrolled participants, 160 were in the HCC group, 88 were in the CHB group and 45 were in the HC group. Baseline characteristics of the enrolled participants were presented at Table 2.
Methylation status of serum TGR5 promoter in different group
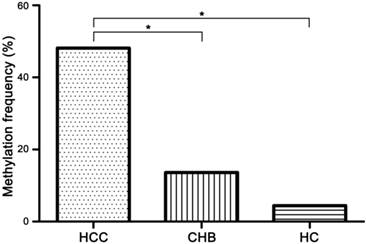
Methylation status of TGR5 promoter was detected in serum of all participants (Figure 2). 77 of 160 (48.13%) HCC patients, 12 of 88 (13.64%) CHB patients and 2 of 45 (4.44%) HCs exhibited TGR5 promoter methylation. The methylation frequency in HCC patients was significantly higher than that in CHB patients (χ2=29.35, p<0.01) and HCs (χ2=28.29, p<0.01). There was no significant difference between the methylation frequency of CHB patients and HCs (χ2=2.67, p=0.10) (Figure 3).
Within the 33 CHB patients with cirrhosis, 6 patients (18.18%) exhibited TGR5 promoter methylation. Meanwhile, within the 55 CHB patients without cirrhosis, 6 patients (10.91%) exhibited TGR5 promoter methylation. No significant difference (p=0.354) was found between the methylation frequency of CHB patients with or without cirrhosis.
Flow diagram depicting the participants' selection process
Characteristics of the enrolled participants at baseline
| Variable | HCC group (n=160) | CHB group (n=88) | HC group (n=45) |
|---|---|---|---|
| Age (yr) | 55.00(46.25-61.00) | 46.50(39.25-58.00) | 42.00(37.5-54.50) |
| Male gender (%) | 132(82.5%) | 61(69.32%) | 28(62.22%) |
| HBeAg+, N(%) | 35(21.88%) | 46(52.27%) | NA |
| ALT (U/L) | 34.00(21.25-66.00) | 87.5 (35.00-155.00) | NA |
| AST (U/L) | 40.50(25.25-81.00) | 66.00(44.50-116.50) | NA |
| TBIL (μmol/L) | 15.70 (11.85-22.70) | 22.75 (14.53-49.58) | NA |
| ALB (g/L) | 40.00(35.00-42.88) | 34.00(28.68-41.70) | NA |
| PT-INR | 1.03(0.97-1.09) | 1.14(1.00-1.34) | NA |
| AFP (ng/mL) | 60.22(4.36-276.70) | 22.05(4.13-50.64) | NA |
| Methylation, N(%) | 77(48.13%) | 12(13.64%) | 2(4.44%) |
NA, not available
Typical MSP analysis results of TGR5 gene promoter. M, methylated sequence; U unmethylated sequence; HCC, hepatocellular carcinoma; CHB, chronic hepatitis B; HC, healthy control; PC, positive control; NC, negative control; WB, water blank.
The methylation frequency of TGR5 promoter in serum of hepatocellular carcinoma (HCC), chronic hepatitis B (CHB) and healthy control (HC) group. 77 of 160 (48.13%) HCC patients, 12 of 88 (13.64%) CHB patients and 2 of 45 (4.44%) HCs exhibited aberrant TGR5 promoter methylation. * significant difference (P<0.05).
Correlation between TGR5 promoter methylation and hepatocellular carcinoma
The TGR5 hypermethylation was more frequently observed in HCCs patients with age ≥60yr (χ2=5.51, p=0.019; Table 3) than those <60yr. TGR5 methylation was not related to gender, HBeAg, smoking, alcohol, primary tumor number, tumor size, histological grading, venous invasion, Tumor Node Metastasis (TNM) staging, Okuda staging and Child-Turcotte-Pugh (CTP) staging (Table 3). No correlation was found between TGR5 hypermethylation and serum AFP level (p=0.638) in HCC and CHB patients.
The diagnostic value of combination of TGR5 promoter methylation and AFP in HCC
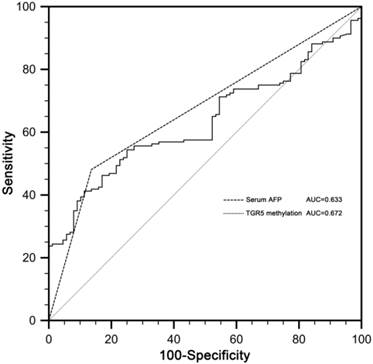
Receiver operating characteristic (ROC) curves for serum AFP and TGR5 promoter methylation in discriminating HCC from CHB patients were constructed. The area under the ROC curves (AUC) of TGR5 methylation in discriminating HCC from CHB patients was 0.672 (standard error [SE] 0.027, 95% confidence interval [CI] 0.610-0.730). The AUC of serum AFP was 0.633 (SE 0.035, CI 0.569-0.693). There was no significant difference between AUC of serum AFP and TGR5 promoter methylation (p=0.388) (Figure 4).
The receiver operating characteristic (ROC) curves of serum AFP and TGR5 methylation in discriminating HCC from CHB patients. The area under the ROC curves (AUC) of TGR5 methylation was 0.672 (standard error [SE] 0.027, 95% confidence interval [CI] 0.610-0.730). The AUC of AFP was 0.633 (SE 0.035, CI 0.569-0.693).
When used to discriminate HCC from CHB patients, aberrant serum TGR5 methylation showed a sensitivity of 48.13% (77/160), specificity of 86.36% (76/88) and Youden index of 0.34. The diagnostic accuracy of AFP and TGR5 methylation combined AFP in discriminating HCC from CHB patients was assessed at AFP cut-off points of 20, 200 and 400ng/ml respectively. At cut-off point of 20ng/ml, AFP had a sensitivity of 58.18%, specificity of 47.73% and Youden index of 0.06; while TGR5 methylation combined AFP had a sensitivity of 81.25%, specificity of 38.64% and Youden index of 0.20. At cut-off point of 200ng/ml, AFP had a sensitivity of 30.63%, specificity of 92.05% and Youden index of 0.23; while TGR5 methylation combined AFP had a sensitivity of 68.13%, specificity of 78.41% and Youden index of 0.47. At cut-off point of 400ng/ml, AFP had a sensitivity of 24.38%, specificity of 98.86% and Youden index of 0.23; while TGR5 methylation combined AFP had a sensitivity of 65.00%, specificity of 85.23% and Youden index of 0.50 (Table 4).
Discussion
In our study, we investigated the serum DNA methylation for TGR5 gene promoter in 160 Patients with HCC, 88 patients with CHB and 45 HCs. Our result showed that the methylation frequency was significantly higher in HCC patients than CHB patients and HCs. The methylation rate of TGR5 in HCC patients ≥60 years old was significantly higher than those <60 years old (p<0.05). When combined with TGR5 methylation, the diagnostic value of AFP for discriminating HCC from CHB was improved at cut-off points of 20, 200 and 400ng/ml respectively.
Correlation between TGR5 methylation status and clinicopathological parameters in HCC patients
| TGR5 methylation status | ||||
|---|---|---|---|---|
| Parameters | Total number | Methylated (%) | Unmethylated (%) | P value |
| Gender | 0.827 | |||
| Male | 132 | 63(48%) | 69(52%) | |
| Female | 28 | 14(50%) | 14(50%) | |
| Age (yr) | 0.019* | |||
| ≤60 | 106 | 44(42%) | 62(58%) | |
| >60 | 54 | 33(61%) | 21(39%) | |
| HBeAg | 0.747 | |||
| negative | 125 | 61(49%) | 64(51%) | |
| positive | 35 | 16(46%) | 19(54%) | |
| Smoking | 0.893 | |||
| No | 84 | 40(48%) | 44(52%) | |
| Yes | 76 | 37(49%) | 39(51%) | |
| Alcohol | 0.908 | |||
| No | 99 | 48(48%) | 51(51%) | |
| Yes | 61 | 29(48%) | 32(52%) | |
| Primary tumor number | 0.571 | |||
| single | 94 | 47(50%) | 47(50%) | |
| multiple | 66 | 30(45%) | 36(55%) | |
| Tumor size | 0.494 | |||
| ≤3cm | 52 | 23(44%) | 29(56%) | |
| >3cm | 108 | 54(50%) | 54(50%) | |
| Histological grading | 0.458 | |||
| poor | 48 | 22(46%) | 26(54%) | |
| moderate | 83 | 38(46%) | 45(54%) | |
| well | 29 | 17(59%) | 12(41%) | |
| Venous invasion | 0.765 | |||
| negative | 83 | 39(47%) | 44(53%) | |
| positive | 77 | 38(49%) | 39(51%) | |
| TNM staging | 0.472 | |||
| I/II | 94 | 43(46%) | 51(54%) | |
| III/IV | 66 | 34(52%) | 32(48%) | |
| Okuda staging | 0.225 | |||
| I/II | 144 | 67(47%) | 77(53%) | |
| III | 16 | 10(63%) | 6(37%) | |
| CTP staging | 0.354 | |||
| A/B | 154 | 73(47%) | 81(53%) | |
| C | 6 | 4(67%) | 2(33%) | |
*, significant difference (P<0.05); TNM, Tumor Node Metastasis; CTP, Child-Turcotte-Pugh
Diagnostic value of AFP and TGR5 methylation combined AFP for discriminating HCC from CHB patients at different cut-off points
| Model | Cut-off points of AFP (ng/mL) | All patients | CHB patients | HCC patients | Sensitivity (%) | Specificity (%) | Youden index |
|---|---|---|---|---|---|---|---|
| AFP alone | n=248 | n=88 | n=160 | ||||
| ≤20 | 109 | 42 | 67 | ||||
| >20 | 139 | 46 | 93 | 58.13 | 47.73 | 0.06 | |
| ≤200 | 192 | 81 | 111 | ||||
| >200 | 56 | 7 | 49 | 30.63 | 92.05 | 0.23 | |
| ≤400 | 208 | 87 | 121 | ||||
| >400 | 40 | 1 | 39 | 24.38 | 98.86 | 0.23 | |
| TGR5 combined AFP | n=248 | n=88 | n=160 | ||||
| ≤20 | 64 | 34 | 30 | ||||
| >20 | 184 | 54 | 130 | 81.25 | 38.64 | 0.20 | |
| ≤200 | 120 | 69 | 51 | ||||
| >200 | 128 | 19 | 109 | 68.13 | 78.41 | 0.47 | |
| ≤400 | 131 | 75 | 56 | ||||
| >400 | 117 | 13 | 104 | 65.00 | 85.23 | 0.50 |
Hepatocellular carcinoma (HCC) is one of the most common and rapidly fatal human malignancies. Most HCC are diagnosed at an advanced stage and died within one year of diagnosis due to the lack of effective surveillance alternatives [25]. Currently available surveillance tests mainly included AFP analysis and ultrasound. However, the effectiveness of them remains controversial [26]. Hypermethylation in promoter regions is now recognized as an important early event in carcinogenesis and detection of methylated DNA has been suggested as a potential biomarker for early detection of cancer. Until now, many genes including GSTP1, RASSF1A, APC, and P16 were found to be aberrantly methylated in serum or tissue of patients with HCC and may potentially be used as biomarkers [11-14].
TGR5, which is a membrane-bound receptor for bile acids, had been demonstrated to be involved in liver carcinogenesis. Our study for the first time revealed that TGR5 promoter aberrant hypermethylation existed in the serum of HCC patients. 77 of 160 (48.13%) HCC patients exhibited aberrant TGR5 promoter methylation in our study, which was significantly higher than CHB patients (12/88, 13.64%) and HCs (2/45, 4.44%). This finding implied that the methylated TGR5 gene in serum may serve as a new biomarker for the surveillance of HCC.
Our study found that the TGR5 hypermethylation was more frequently observed in HCCs patients ≥60yr (χ2=5.51, p=0.019) than those <60yr, which is consistent with several other studies. Xin Li et al. showed that hypermethylation of the p16INK4A-promoter region was more frequent in HCCs from older patients [27]. Ahuja N et al. determined the frequency of age-related methylation in normal colonic mucosa among the genes hypermethylated in colorectal cancer and suggested that aging is a major contributing factor to hypermethylation in cancer [28]. However, the study performed by Masahito Tsutsui et al. showed that methylation of CCND2 gene was not related to age [29]. It is possible that age-related methylation may be gene specific and tumor specific.
CHB infection is a major risk factor for HCC worldwide. Beasley et al., in a prospective controlled study showed that the annual incidence of HCC in hepatitis B carriers was 0.5% [30]. In patients with known CHB related cirrhosis, the incidence was 2.5%/year. CHB patients are recommended to be screened at 6 to12 month intervals. Until now, the screening and surveillance for HCC mainly depended on serum level of alpha fetoprotein (AFP) and ultrasonography. AFP alone is not recommended for screening unless ultrasound is not available because of a relatively low diagnostic value and accuracy [31]. The most widely used cut-off points of AFP are 20, 200 and 400ng/mL. In our study, the sensitivity of AFP increased from 58.13% (93/160) to 81.25% (130/160) at cut-off point of 20ng/ml with the combination of which meant AFP could reveal 37 more HCC patients when combined with TGR5 methylation. The sensitivity of AFP increased from 30.63% (49/160) to 68.13% (109/160) at cut-off point of 200ng/ml with the combination of TGR5 methylation, which meant AFP could reveal 60 more HCC patients when combined with TGR5 methylation. The sensitivity of AFP increased from 24.38% (39/160) to 65.00% (104/160) at cut-off point of 400ng/ml with the combination of TGR5 methylation, which meant AFP could reveal 65 more HCC patients when combined with TGR5 methylation. Although lack adequate sensitivity and specificity, AFP was still the most widely used serum biomarker for screening HCC until now [31]. Our study compared the combination of AFP and TGR5 methylation with AFP alone in discriminating HCC from CHB patients and showed that the combination of AFP and TGR5 methylation could enhance the diagnostic value of AFP at different cut-off points. The comparison between the combination of TGR5 methylation and AFP with other HCC markers could be done in our further research.
However, there are some limitations in this present study. First, we didn't analyze the methylation status and mRNA expression of TGR5 gene in HCC tissue or HCC cell lines. DNA methylation is one of the most important epigenetic mechanisms and methylation of gene promoter regions links to long-term stabilization of transcriptional silencing and loss of gene function [32-35]. Of great importance, increasing clinical evidences have shown that serum DNA methylation of some genes could be recognized as novel biomarkers for the diagnosis of human cancers including prostates cancers, gastric cancers, liver cancers and so on [36-39]. In the patients with hepatocellular carcinoma, we have previously demonstrated serum DNA methylation of tissue factor pathway inhibitor-2 promoter could be identified as a potential biomarker for HCC [11]. In our present study, we therefore mainly intended to find the possible diagnostic value of TGR5 promoter methylation as a novel biomarker for HCC patients. Obtained results suggested combination of serum TGR5 promoter methylation and AFP might enhance the diagnostic value of AFP alone in discriminating HCC from CHB patients. However, we believe that these results should be carefully explained in multiple aspects especially in the mechanism manner of biological function analysis for TGR5 promoter methylation. Experimental evidences using liver tissue and HCC cell lines should be done in our further study to demonstrate the exact function of TGR5 promoter methylation on the regulation of TGR5 expression and the possible role of TGR5 promoter methylation in the development of HCC. Second, we did not confirm our study in a second cohort. The validation test of our finding using second cohort and well-matched cohort should be performed in our further research. Third, we only enrolled HBV-infected HCC patients and revealed that the combination of serum TGR5 promoter methylation and AFP enhanced the diagnostic value of AFP alone in discriminating HCC from CHB patients. However, the “tumor suppressor” effect of Gpbar1 should not be restricted in HBV infected HCC patients and there are also some other kinds HCC patients with different causes including HCV and alcoholism. Therefore, we believed that the other HCC patients with HCC and alcoholism should be performed in our further research
In conclusion, our study demonstrated that the aberrant hypermethylation of TGR5 promoter in serum cfDNA might serve as a biomarker for the surveillance of HCC. TGR5 hypermethylation was significantly associated with aging. It could enhance the diagnostic value of AFP in discriminating HCC from CHB patients at cut-off points of 20, 200 and 400ng/mL respectively.
Supplementary Material
Supplementary Figure 1Abbreviations
TGR5: G-protein-coupled bile acid receptor Gpbar1; HCC: hepatocellular carcinoma; CHB: chronic hepatitis B; HC: healthy control; AFP: alpha fetoprotein; cfDNA: circulating cell-free DNA; HBV: hepatitis B virus; MSP: methylation-specific polymerase chain reaction; NF-κB: nuclear factor kappa light-chain enhancer of activated B cells
Acknowledgements
This work was supported by grants from Key Project of Chinese Ministry of Science and Technology (2012ZX10002007, 2013ZX10002001), and National Natural Science Foundation of China (81171579, 81201287, 81371832).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
2. Perz JF, Armstrong GL, Farrington LA. et al. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538
3. Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907-1917
4. El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745-750
5. Deuffic S, Poynard T, Buffat L. et al. Trends in primary liver cancer. Lancet. 1998;351:214-215
6. El-Serag HB, Marrero JA, Rudolph L. et al. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752-1763
7. Cokus SJ, Feng S, Zhang X. et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215-219
8. Oshimo Y, Nakayama H, Ito R. et al. Promoter methylation of cyclin D2 gene in gastric carcinoma. Int J Oncol. 2003;23:1663-1670
9. Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-116
10. Chan TA, Baylin SB. Epigenetic biomarkers. Curr Top Microbiol Immunol. 2012;355:189-216
11. Sun FK, Fan YC, Zhao J. et al. Detection of TFPI2 Methylation in the Serum of Hepatocellular Carcinoma Patients. Dig Dis Sci. 2012 [Epub ahead of print]
12. Huang ZH, Hu Y, Hua D. et al. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp MolPathol. 2011;91:702-707
13. Zhang YJ, Wu HC, Shen J. et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13:2378-2384
14. Wang J, Qin Y, Li B. et al. Detection of aberrant promoter methylation of GSTP1 in the tumor and serum of Chinese human primary hepatocellular carcinoma patients. Clin Biochem. 2006;39:344-348
15. Watanabe M, Houten SM, Mataki C. et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484-489
16. Maruyama T, Tanaka K, Suzuki J. et al. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol. 2006;191:197-205
17. Thomas C, Gioiello A, Noriega L. et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167-177
18. Wang YD, Chen WD, Yu D. et al. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology. 2011;54:1421-32
19. Chen WD, Yu D, Forman BM. et al. Deficiency of G-protein-coupled bile acid receptor Gpbar1 (TGR5) enhances chemically induced liver carcinogenesis. Hepatology. 2013;57:656-66
20. Akizu N, Estarás C, Guerrero L. et al. H3K27me3 regulates BMP activity in developing spinal cord. Development. 2010;137:2915-25
21. Li C, Chu N, Wang B. et al. Polymorphisms in the presumptive promoter region of the SLC2A9 gene are associated with gout in a Chinese male population. PLoS One. 2012;7:e24561
22. Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022
23. Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661-662
24. Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427-31
25. Schafer DF, Sorrell MF. Hepatocellular carcinoma. Lancet. 1999;353:1253-7
26. De Masi S, Tosti ME, Mele A. Screening for hepatocellular carcinoma. Dig Liver Dis. 2005;37:260-8
27. Li X, Hui AM, Sun L. et al. p16INK4A hypermethylation is associated with hepatitis virus infection, age, and gender in hepatocellular carcinoma. Clin Cancer Res. 2004;10:7484-9
28. Ahuja N, Li Q, Mohan AL. et al. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489-94
29. Tsutsui M, Iizuka N, Moribe T. et al. Methylated cyclin D2 gene circulating in the blood as a prognosis predictor of hepatocellular carcinoma. Clin Chim Acta. 2010;411:516-20
30. Beasley RP, Hwang LY, Lin CC. et al. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129-1133
31. Jordi Bruix, Morris Sherman. Management of Hepatocellular Carcinoma. Hepatology. 2005;42:1208-36
32. Irizarry RA, Ladd-Acosta C, Wen B. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178-186
33. Doi A, Park IH, Wen B. et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350-1353
34. Ji H, Ehrlich LI, Seita J. et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338-342
35. Futscher BW, Oshiro MM, Wozniak RJ. et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175-179
36. Liu L, Kron KJ, Pethe VV. et al. Association of tissue promoter methylation levels of APC, TGFbeta2, HOXD3 and RASSF1A with prostate cancer progression. Int J Cancer. 2011;129:2454-2462
37. Takamaru H, Yamamoto E, Suzuki H. et al. Aberrant methylation of RASGRF1 is associated with an epigenetic field defect and increased risk of gastric cancer. Cancer Prev Res. 2012;5:1203-12
38. Lambert MP, Paliwal A, Vaissière T. et al. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J Hepatol. 2011;54:705-15
39. Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22:50-5
Author contact
![]() Corresponding author: Kai Wang, MD, PhD, Department of Hepatology, Qilu Hospital of Shandong, University and Hepatology Institute of Shandong University, Jinan 250012, China, Wenhuaxi Road 107#, Jinan 250012, Shandong, China. E-mail: wangdoc876com; wangdoc2010com; Tel: +86-531-86630809; Fax: +86-531-86927544
Corresponding author: Kai Wang, MD, PhD, Department of Hepatology, Qilu Hospital of Shandong, University and Hepatology Institute of Shandong University, Jinan 250012, China, Wenhuaxi Road 107#, Jinan 250012, Shandong, China. E-mail: wangdoc876com; wangdoc2010com; Tel: +86-531-86630809; Fax: +86-531-86927544

 Global reach, higher impact
Global reach, higher impact