Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(10):1333-1343. doi:10.7150/ijms.5774 This issue Cite
Research Paper
Allele Dependent Silencing of Collagen Type I Using Small Interfering RNAs Targeting 3'UTR Indels - a Novel Therapeutic Approach in Osteogenesis Imperfecta
1. Department of Medical Sciences, Uppsala University, Uppsala, Sweden
2. Neuropediatric unit, Karolinska University Hospital and Department of Women's and Children's Health, Karolinska Institutet, Stockholm, Sweden.
3. Department of Medical Biochemistry and Microbiology, Uppsala University, Sweden
Received 2012-12-28; Accepted 2013-7-23; Published 2013-8-13
Abstract
Osteogenesis imperfecta, also known as “brittle bone disease”, is a heterogeneous disorder of connective tissue generally caused by dominant mutations in the genes COL1A1 and COL1A2, encoding the α1 and α2 chains of type I (pro)collagen. Symptomatic patients are usually prescribed bisphosphonates, but this treatment is neither curative nor sufficient. A promising field is gene silencing through RNA interference. In this study small interfering RNAs (siRNAs) were designed to target each allele of 3'UTR insertion/deletion polymorphisms (indels) in COL1A1 (rs3840870) and COL1A2 (rs3917). For both indels, the frequency of heterozygous individuals was determined to be approximately 50% in Swedish cohorts of healthy controls as well as in patients with osteogenesis imperfecta. Cultures of primary human bone derived cells were transfected with siRNAs through magnet-assisted transfection. cDNA from transfected cells was sequenced in order to measure targeted allele/non-targeted allele ratios and the overall degree of silencing was assessed by quantitative PCR. Successful allele dependent silencing was observed, with promising results for siRNAs complementary to both the insertion and non-insertion harboring alleles. In COL1A1 cDNA the indel allele ratios were shifted from 1 to 0.09 and 0.19 for the insertion and non-insertion allele respectively while the equivalent resulting ratios for COL1A2 were 0.05 and 0.01. Reductions in mRNA abundance were also demonstrated; in cells treated with siRNAs targeting the COL1A1 alleles the average COL1A1 mRNA levels were reduced 65% and 78% compared to negative control levels and in cells treated with COL1A2 siRNAs the average COL1A2 mRNA levels were decreased 26% and 49% of those observed in the corresponding negative controls. In conclusion, allele dependent silencing of collagen type I utilizing 3'UTR indels common in the general population constitutes a promising mutation independent therapeutic approach for osteogenesis imperfecta.
Keywords: osteogenesis imperfecta, collagen type I, siRNA
Introduction
Osteogenesis imperfecta (OI) is a clinically and genetically heterogeneous disease of connective tissue with an incidence of approximately 1/10-20 000. The principal symptom is an increased susceptibility to fractures, with the most severe forms being perinatally lethal, often due to extreme bone fragility with pulmonary hypoplasia and respiratory insufficiency. OI is traditionally divided into four types based on clinical phenotype, where the mildest form (type I) is usually due to a quantitative collagen type I defect[1-3], while severe and lethal forms (types II-III and severe type IV) have qualitative collagen defects[2-4]. Approximately 90% of OI is the result of dominantly inherited mutations in collagen type I, which is the most abundant protein in connective tissue. However, to date nine rare recessive gene variants[5], accounting for approximately 5-10% of clinical OI, have been discovered and the genetics of the disease are thus more complex than initially believed. Recessive loci have so far been found in genes regulating osteoblastogenesis and affecting collagen assembly and processing[5].
The organic matrix in skeletal tissue consists primarily of collagen type I, which provides both the framework for mineralization and the tensile strength that gives bone resilience to torsion and bending powers. The collagen type I heterotrimer is composed of two α1(I) chains and one α2(I) chain, encoded by the genes COL1A1 and COL1A2 respectively. The three monomers twist together in a zipper-like fashion to create a triple helix with a highly repetitive structure of glycine-X-Y triplet repeats, glycine being the only amino acid small enough to sterically fit in the confined center of the helix. A mutation of a helical glycine residue is believed to interfere with proper helix formation resulting in collagen over-modification and accounts for approximately 80% of structurally abnormal collagen in OI, while splice site mutations constitute around 20% of qualitative mutations[4].
Bisphosphonates are the drug of choice for symptomatic treatment of patients with OI [6,7], but their effect is neither sufficient nor satisfactory. Treatment with other osteoporosis pharmaceuticals could be a more beneficial alternative or perhaps a complement to bisphosphonates in patients with OI, but the knowledge of which patients benefit from certain treatment regimens is limited. Considering that severe OI commonly is a dominantly inherited disease, an attractive novel approach is gene silencing of the mutated allele. Successful allele preferential silencing would in theory convert a severe OI to an OI type I (null allele) in the case of a COL1A1 mutation and to a phenotypically normal individual in the case of a COL1A2 mutation[8]. And indeed, the first steps toward allele dependent silencing have been achieved using small interfering RNAs (siRNAs)[9,10].
Cellular RNA exposed to complementary RNA can be degraded in a process called RNA interference (RNAi). This method of inhibition is very specific and has a high inhibitory activity [11]. RNAi mediated by endogenous siRNAs in plants was discovered in 1999 [12] and in 2001 it was shown in vitro that exogenous double stranded siRNAs could reduce mRNA levels efficiently in animal cells[13]. Since these original discoveries the rules determining successful mRNA suppression by siRNAs have been extensively studied and siRNAs are now invaluable tools for studies of partial gene knockout in vitro as well as in vivo[14]. Studies of siRNAs that discriminate between single nucleotide variants within mRNAs have shown that allele specific silencing utilizing siRNA technology is possible[15-20]. This suggests that siRNAs may be a way forward in the treatment of dominant monogenetic diseases, such as OI. Allele discriminatory inhibition of COL1A1 mediated by siRNAs has been successfully achieved in human mesenchymal progenitor cells[9] as well as in human bone derived cells, the latter by targeting a common disease-unrelated exonic SNP in COL1A2 [10].
More than 800 mutations in COL1A1 and COL1A2 that cause OI through structural collagen changes have been described[4] and new mutations are frequently reported. Hence creating unique siRNAs, each targeting one mutation would be labor intensive. A possible mutation independent approach is designing siRNAs targeting common COL1A1 and COL1A2 SNPs, [9] and this approach has been pursued by the authors previously[10]. However, satisfactory allele specificity was difficult to achieve considering that the two alleles differ by only one nucleotide in a heterozygous individual and COL1A1 and COL1A2 are very repetitive genes. Furthermore, there are virtually no common exonic SNPs in COL1A1 to target, however a common insertion/deletion polymorphism (indel) was noted in the 3'untranslated regions (3'-UTR) of both COL1A1 and COL1A2. Utilizing siRNAs targeting heterozygotes for indels is a theoretically attractive approach as in this case the two alleles differ by more than one nucleotide, which should increase the chance of true allele-specificity.
In this study the allele discriminatory effects of siRNAs targeting two 3'-UTR indels in COL1A1 (rs38408701) and COL1A2 (rs3917) have been studied in primary human bone derived cells obtained from individuals with heterozygote genotypes for the indels. Both collagen type I indels have close to 50% heterozygote allele frequencies in Europeans (Table 1 for heterozygous frequencies in several populations). To our knowledge, this approach has not previously been utilized in allele-preferential silencing of collagen type I.
Frequencies of heterozygotes for rs3840870 and rs3917 in different populations
| Population/ Ethnicity | Contributing Alleles | rs3840870 (COL1A1) Heterozygotes | rs3917 (COL1A2) Heterozygotes |
|---|---|---|---|
| MrOS cohort | 192 | 52% | 46% |
| OI cohort | 192 | 52% | 48% |
| European origin° | 120 | 47% | 49% |
| Youruban Nigeria° | 120 | 50% | 17% |
| Chinese/Japanese° | 120 | 49% | 11% |
| Not stated [35] | 240 | 46% | N/A |
| European* | 200 | N/A | 49% |
| Amerind* | 50 | N/A | 49% |
| Japanese* | 50 | N/A | 35% |
| African* | 42 | N/A | 38% |
°= Data from 1000 Genomes Project (http://www.1000genomes.org/)
*= Data from NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/)
Percentages of individuals with a heterozygous genotype of rs3840870 and rs3917 were genotyped in 96 Swedish healthy controls (subset of the MrOS cohort) and in 96 Swedish patients with OI and results are presented here. Allele frequencies for the indels in the National Centre for Biotechnology Information (NCBI) dbSNP database and the 1000 Genomes Project were extrapolated to heterozygous frequencies using the Hardy-Weinberg equilibrium. The number of contributing alleles is stated for each group and the specified ethnicity as stated in the respective submitting sources.
Materials and methods
Subjects
MrOS cohort
In order to establish the frequencies of heterozygotes for the two indels in the general Swedish population 96 Swedish men from the population based MrOS cohort[21] were genotyped for rs3840870 and rs3917. The Swedish regional ethics committee at Uppsala University (Ups 01-057) approved the study and written consent was acquired from all participants.
Cohort of patients with OI
To establish the frequencies of heterozygotes for the two indels in a population of Swedish OI patients, 96 patients with OI from Astrid Lindgren Children's Hospital, Stockholm and Uppsala University Hospital, Uppsala were genotyped for rs3840870 and rs3917 after obtaining written consent as part of a study approved by the Swedish regional ethics committee at Uppsala University (Ups 06-212). In the case of children under 18 years of age written consent was obtained from parent or legal guardian.
siRNA design
For each of the two indels rs3840870 and rs3917 two double stranded siRNAs were designed, one with an antisense strand perfectly complementary to the allele carrying the insertion and the other with an antisense strand perfectly complementary to the allele not carrying the insertion (Figure 1 illustrates siRNA complementarity) (Table 2 exhibits siRNA names and sequences). The siRNAs were analyzed for complementarity with other targets, but no other 100% matches were discovered. The siRNAs were purchased from Ambion, USA as double stranded RNA molecules. Each strand had a two-base pair overhang in the 3'-end (always UU for sense strand). Negative control siRNAs were purchased from Invitrogen and were: Stealth RNAi ™ siRNA Negative Controls (part numbers: NC1: 12935-200 and NC2: 12935-112).
Four siRNAs were designed to target COL1A1 and COL1A2 3'UTR indels. Four siRNAs were designed to target the region surrounding the four base pair indel rs3840870 in COL1A1 and the seven base pair indel rs3917 in COL1A2. One siRNA was created to target each allele of a heterozygous individual, and a total of four siRNAs were used in the studies. The complementarity of the active anti sense strand is illustrated in relation to collagen 3'UTRs with and without the insertions. Insertion bases are marked red while the blue arrows indicate the insertion location in alleles without insertion.
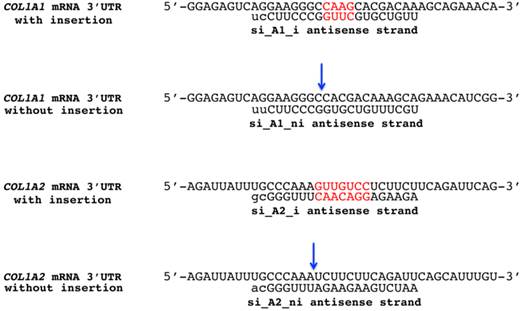
Names and sequences of COL1A1 and COL1A2 3'UTR indel targeting siRNAs
| Name | Target | Sequence | Strand |
|---|---|---|---|
| si_A1_i | COL1A1 allele carrying an insertion | GAAGGGCCAAGCACGACAAuu | Sense strand |
| ucCUUCCCGGUUCGUGCUGUU | Anti sense strand | ||
| si_A1_ni | COL1A1 allele not carrying an insertion | GAAGGGCCACGACAAAGCAuu | Sense strand |
| uuCUUCCCGGUGCUGUUUCGU | Anti sense strand | ||
| si_A2_i | COL1A2 allele carrying an insertion | CCCAAAGUUGUCCUCUUCUuu | Sense strand |
| gcGGGUUUCAACAGGAGAAGA | Anti sense strand | ||
| si_A2_ni | COL1A2 allele not Carrying an insertion | CCCAAAUCUUCUUCAGAUUuu | Sense strand |
| acGGGUUUAGAAGAAGUCUAA | Anti sense strand |
Four siRNAs were designed to target the region surrounding the four base pair indel rs3840870 in COL1A1 and the seven base pair indel rs3917 in COL1A2. One siRNA was created to target each allele of a heterozygous individual. Each strand had a two-nucleotide 3'-overhang, indicated by lower-case letters. Denotation, sense and active anti sense strand for each siRNA are displayed in the table.
Cell culture and transfection
Cell cultures of primary human bone cells were obtained from patients undergoing orthopedic surgery at Uppsala University Hospital (approved by the Swedish regional ethics committee at Uppsala University (Ups 03-561) and written consent was obtained from all patients). The cultures were genotyped and cells obtained from individuals heterozygous for rs3840870 and/or rs3917 were subsequently used in transfection experiments with the four siRNAs. Cells were only submitted to one passage prior to transfection to minimize differentiation from primary human bone cells.
The day prior to transfection 35,000 cells were seeded in 24-well cell culture plates and were cultured at 37°C in the presence of 5% CO2 overnight. According to previous experience this cell count corresponds to manufacturer's recommended 50% maximum density, which is reflected as confluency for primary human bone cells. Transfection was carried out using magnet-assisted transfection for siRNAs (MATra-si) (Promokine, Germany). There is generally a relation between the amount of target mRNA present and silencing siRNA required for optimal silencing with minimum off target effects. The exact ratio for each specific pair of mRNA:siRNA is not known, and our approach entailed a dose-response study and time-course scheme to find optimal dose and time points. Cells were transfected with varying concentrations (0.025µg, 0.05µg, 0.1µg, 0.2µg, 0.4µg and 0.6µg) of each siRNA and also with two separate negative control siRNAs as a pilot study. Selection of the highest concentration was guided by recommendations in the manufacturer's protocol and five successively lower doses were also chosen, ending with 1/24 of the recommended initial trial dose for complete gene silencing. Amount of MATra-si is determined by the dose of siRNA.
Post-transfection, cells were incubated in a humidified chamber for 72 h at 37°C in the presence of 5% CO2 after which RNA was prepared. A time-course experiment was subsequently performed with incubation of cells for 24, 48, 72, 120, 168 and 216 hours after transfection with 0.6µg siRNA for COL1A1-targeting siRNAs and 0.4µg siRNA for COL1A2-targeting siRNAs; doses based on results in the pilot study. Finally a repeated dose response study was performed and RNA was extracted 72 h post transfection, as the allele ratio shift was maximized around 72 hours post transfection according to the time course experiment. Negative controls were transfected with 0.6µg negative control siRNA in all instances except for in the time-course COL1A2 study where 0.4µg was used for the negative controls as well. Each transfection was performed in triplicate. For each gene in each separate experiment cells from one individual were utilized, and over the full set of experiments cells from four donors were used to obtain the results described altogether. SiRNA amount is stated as a dose per well considering that the most relevant information is the amount of siRNA being magnetically drawn in to the adherent cells during the transfection step performed on a magnet, and not the siRNA-magnetic bead complex concentration in the medium above the cell-layer prior to transfection.
RNA preparation and cDNA-synthesis
Total RNA was isolated using the QiaShredder kit and the RNeasy mini kit (Qiagen, Germany). Each individual RNA-sample was subjected to DNase treatment using TURBO-DNAfree (Ambion, USA) and cDNA-synthesis was then carried out using the High Capacity cDNA reverse transcription kit (Applied Biosystems, USA).
Polymerase Chain Reaction and sequencing
The cDNA samples were used as templates in polymerase chain reactions (PCRs) where regions of COL1A1 and COL1A2 mRNA surrounding the polymorphisms rs3840870 and rs3917 were amplified (Table 3 for primers). PCR-reactions were performed using standard PCR conditions with an annealing temperature of 60°C. The sequencing reaction was performed using the same primers with an adjusted Big Dye Terminator 3.1 sequencing protocol (Applied Biosystems, USA). The product was run on a 16-capillary ABI 3130xl Genetic Analyzer automated sequencer (Applied Biosystems, USA).
Allele frequencies of the insertions in the cohorts described above were established using the same procedure with the exception that genomic DNA was sequenced.
Sequences of primers used for PCR and sequencing
| Primer | Sequence |
|---|---|
| rs3840870 forward | 5´GGTTCTCAGACTGCCAAAGAAG |
| rs3840870 reverse | 5´GCAACACAGTTACACAAGGAACA |
| rs3917 forward | 5´TTTCTGCTTGCCCAAGAAAC |
| rs3917 reverse | 5´CTTGAAGAATGTTGATGGTGCT |
Assessment of relative allele abundance of COL1A1 and COL1A2 mRNA
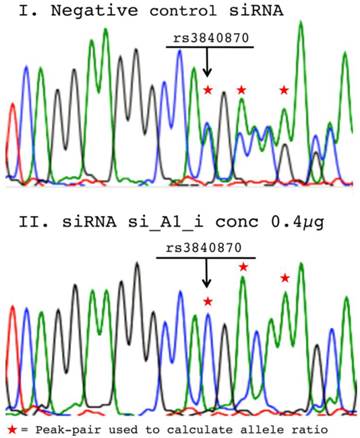
The software PeakPicker [22] was used to determine the relative allele ratios from cDNA-sequencing. Briefly, as Sanger sequences derived from cells heterozygous for short indels were being analyzed, double chromatogram peaks for all positions 3' of the indel were observed in the non-silenced samples. For each individual cDNA-sequence, peak-heights 3' of the insertion were normalized for peak-heights of adjacent non-polymorphic positions. As the sequence after the insertion in a heterozygous individual appears to be predominantly polymorphic, several positions could be compared to the non-polymorphic peaks prior to the insertion. The same three peak-pairs were used for the calculation of the allele ratio in cDNA sequences from siRNA treated and negative control treated cells and an average allele ratio was calculated for cDNA from each well (Figure 2 demonstrates representative sequences and used peak-pairs). For all treatments, allele ratios of the two indels rs3840870 and rs3917 were normalized to peak-heights of corresponding negative controls.
Quantitative PCR
Quantitative PCR reactions were performed using 10µl 2x TaqMan® Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems, USA) mixed with 9 µl diluted cDNA and 1 µl of Taq-man gene specific assay mix. Two probes were used for each collagen gene; COL1A1: Hs01076772_gH and Hs01028970_m1 and COL1A2: Hs01028970_m1 and Hs01028942_gH. The two reference genes glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and beta-actin (ACTB) were used (all probes Applied Biosystems, USA). This mix was subjected to 40 cycles of PCR using the 7500 Fast Real-Time PCR System instrument (Applied Biosystems, USA). Each individual sample was analyzed in triplicate and relative levels of COL1A1 and COL1A2 mRNAs were determined as the average quantity calculated from the two probes used per collagen gene. This quantity was subsequently normalized relative GAPDH and ACTB mRNA levels. The average ratio of collagen vs. reference gene in negative control was defined as 1 and the ratios of collagen in the silenced samples were calculated relative to negative controls.
Chromatograms from negative control vs. siRNA treated cell populations. Representative chromatograms from sequencing of cDNA samples derived from RNA isolated 72h post-transfection with: (I) 0.6µg negative control siRNA and (II) 0.4µg si_A1_i from the time-course study. Arrows mark the position for the COL1A1 rs3840780 insertion and the asterisks mark the peak-pairs used to determine allele ratios using PeakPicker software.
Cy3 labeling of siRNAs to determine transfection efficiency
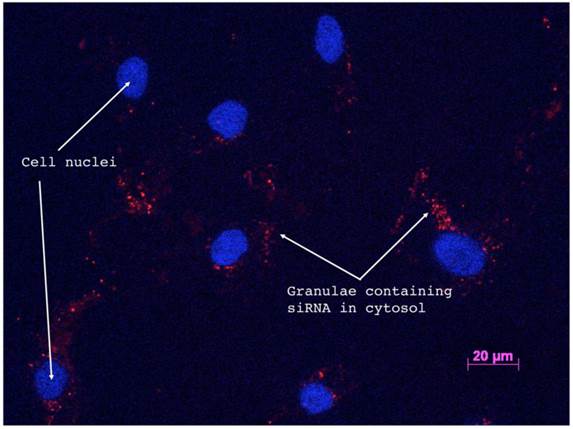
To verify the delivery of siRNAs to the cytosol of cells, and to determine the transfection efficiency the Silencer® siRNA Labeling Kit from Invitrogene was used to label si_A1_i with Cy3. Six wells of primary bone cells were subsequently transfected corresponding to the 0.6µg dose, as this was the highest dose used in the dose-response and time-course experiments. Transfection efficiency was determined by ocular examination of photographs taken with a fluorescent light microscope where visible granulae in the cytosol counted as a positive cell (Figure 3). Three investigators separately calculated the frequency of positive cells and the total number of cells in two representative photographs from each of the six wells and the average efficiency was then calculated. The actual efficiency of each individual siRNA was assessed by monitoring the end result (i.e. allele ratio and quantitative sequencing), rather than determining up-take of cy3-labeled siRNA for all concentrations.
Cell proliferation assay
MG-63 cells (osteosarcoma derived cell line) were seeded in 24-well culture plates at a concentration of 35,000 cells/well, and cultured for 24 hours in a humidified chamber at 37°C in the presence of 5% CO2. Transfection was carried out under identical conditions described above with 0.6 µg COL1A1 or COL1A2 indel targeting siRNA, negative control siRNAs or MATra-si particles only. Post-transfection, cells were incubated for 72 hours at 37°C, after which the level of cell respiration was evaluated by MTS assay, using CellTiter 96®AQueousOne Solution Cell Proliferation Assay (Promega, USA) according to the manufacturer's protocol. Each transfection was performed in 30 replicates. The experiments were performed on this cell line as primary human bone cell cultures have a slow growth rate and there was a need for repeated experiments.
Results
In the 96-subject subset of the MrOS cohort of healthy Swedish control individuals 52% were heterozygous for the 4-nucleotide long COL1A1 indel (rs3840870) and 46% were heterozygous for the 7-nucleotide long COL1A2 indel (rs3917) (Table 1). The frequencies in a cohort of 96 Swedish OI patients were quite similar with 52% heterozygotes for rs3840870 and 48% heterozygotes for rs3917. This suggests that in this population approximately 50% of patients with a dominant collagen type I mutation could be candidates for treatment with allele specific siRNAs targeting these two indels. For allele frequencies in several populations see table 1.
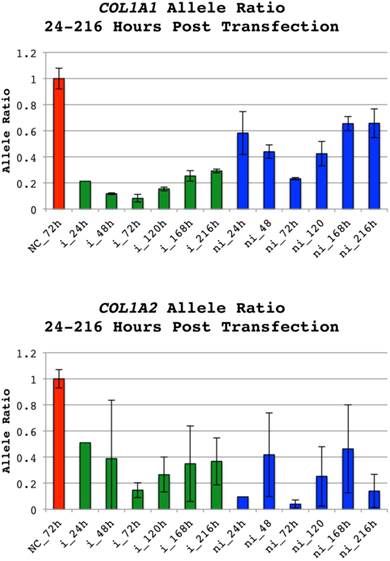
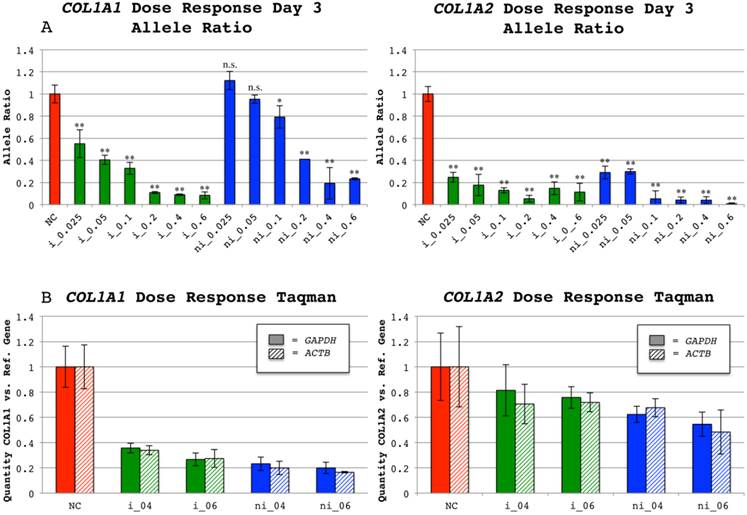
Allele ratios (insertion vs. non-insertion) of negative controls were equal to allele ratios of untreated cells (data not shown) and consequently defined as 1. Successful allele preferential silencing was achieved for all studied collagen siRNAs in a dose dependent manner. In the pilot-study an allele ratio of 0.06 was observed for both siRNAs targeting the insertion harboring alleles while the non-insertion containing alleles were silenced to allele ratios of 0.24 and 0.08 for COL1A1 and COL1A2 respectively (data not shown). The time course study subsequently performed demonstrated that the allele ratio shift was maximized around 72 hours post transfection (Figure 4), although an effect could be seen for all siRNAs from first to last time-point. In the following dose response study the 0.4µg dose of siRNAs targeting COL1A1 shifted the allele ratio from 1 to 0.09 for of si_A1_i and 0.19 for si_A1_ni (Figure 5A). The COL1A2 targeting si_A2_i rendered an allele ratio of 0.05 for the 0.2µg dose and 0.11 for the dose 0.6µg, which was used for subsequent quantitative PCR. An allele ratio of 0.01 was observed for the 0.6µg dose of si_A2_ni (Figure 5A).
Quantitative PCR was used to measure levels of COL1A1 and COL1A2 mRNAs for the 0.4 and 0.6µg doses in the dose response study. The collagen mRNA levels were normalized to levels of housekeeping gene mRNAs (GAPDH and ACTB). Collagen decrease was subsequently presented relative to collagen mRNA levels in negative controls. The expected mRNA abundances can be calculated with the formula 0.5+(0.5 x allele ratio), which assumes that the observed silencing is 100% allele specific and has no effect on mRNA quantity from the non-targeted allele. According to this, the allele ratio of 0.09 for si_A1_i would render an mRNA abundance of 55%, which is equivalent to 45% suppression of normal levels. The 0.4µg doses of si_A1_i and si_A1_ni suppressed the mRNA levels by 65% and 78% compared to the expected 45% and 40% according to allele ratios, presuming perfect allele discrimination described above. Under the same postulate, the 0.6µg dose of si_A2_i and si_A2_ni should decrease mRNA levels by 44% and 49% respectively based on quantitative sequencing, while the mRNA abundances measured by TaqMan were decreased by 26% and 49%, respectively (Figure 5B).
Successful siRNA delivery to the target cells is shown in Figure 3, which depicts a fluorescence microscopy image of the Cy3 labeled si_A1_i transfected cells 72 hours post-transfection. Calculations of positive cells was performed by three investigators who in 12 representative images counted an average of 189 cells out of which 180 were determined to be positive for siRNA uptake, yielding a transfection efficiency of 95%.
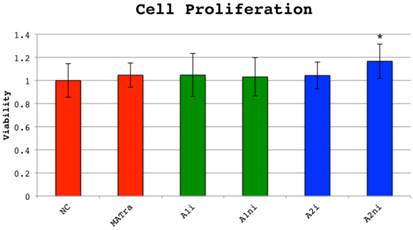
Cell proliferation measured by MTS-assay was not decreased in wells treated with indel targeting siRNAs compared to wells treated with negative control siRNAs or MATra-si only and thus siRNAs did not have an inverse effect on cellular viability. However, si_A2_ni treated MG63 cells had a slightly increased proliferation of unknown significance (Figure 6).
Fluorescence microscopy of Cy3-labled si_A1_i siRNAs confirms cellular uptake. Fluorescence microscopy image of Cy3-labeled si_A1_i inside of primary bone cells 72 h post-transfection. Red staining indicates areas where siRNAs are present and blue regions mark DAPI-stained cellular nuclei.
Allele ratios of COL1A1 and COL1A2 indels 24 to 216 hours post transfection. Allele ratios of insertion(i):non-insertion(ni) alleles for cell populations treated with siRNAs targeting either allele of COL1A1 indel rs3840870 or COL1A2 rs3917. The doses 0.6µg and 0.4µg were used for the COL1A1 and COL1A2 indels and corresponding negative controls (NC) respectively. RNA was prepared 24, 48, 72, 120, 168 and 216 hours after transfection. The software PeakPicker was used to calculate peak heights of cDNA chromatograms. Peak heights were normalized to cells treated with negative control siRNA, which were defined as 1. Error bars indicate standard deviation. The allele ratios for non-insertion treated siRNAs have been inverted to enable presentation of results in the same chart. Color of bars denotes category of siRNA (NC-red, i-green and ni-blue).
Dose response allele ratios and relative mRNA levels of COL1A1 and COL1A2 indels at 72 hours. (A) Ratio of insertion(i)/non-insertion(ni) alleles for cell populations treated with either of the siRNAs targeting COL1A1 indel rs3840870 or COL1A2 indel rs3917 72 hours post transfection. A range of siRNA concentrations was used: 0.025µg, 0.05µg 0.1µg, 0.2µg, 0.4µg and 0.6µg for indel-targeting siRNAs and for negative controls (NC) 0.6µg was used. The software PeakPicker was used to calculate the heights of the same three peak-pairs in all cDNA chromatograms and values were normalized to negative control treated cells, with the mean defined as 1. Error bars indicate standard deviation. The allele ratios for non-insertion treated siRNAs have been inverted to enable presentation of results in the same chart. Color of bars denotes category of siRNA (NC-red, i-green and ni-blue). A two-tailed T-test comparing negative controls with each type and dose of siRNA was performed. * = p < 0.05; ** = p < 0.001 and n.s. = not significant. (B) Average COL1A1 and COL1A2 mRNA levels calculated from the two probes used per collagen gene in relation to levels of control genes (GAPDH and ACTB) measured by quantitative PCR in negative controls (NC) and siRNA treated wells. The mean ratio of COL1A and COL1A2 to either reference gene in negative control wells was defined as 1. Error bars indicate standard deviation. Pattern of bars denote reference gene (solid for GAPDH and striped for ACTB) used in quantitative PCR and color denotes the category of used siRNA (NC-red, i-green and ni-blue).
Cell proliferation 72 hours post-transfection measured by MTS-assay. MG-63 cells (osteosarcoma derived cell line) were transfected with 0.6 µg COL1A1 or COL1A2 indel targeting siRNA, negative control siRNA (NC) or MATra-si particles only. Cell respiration was evaluated by MTS assay after incubation for 72 hours. Cell proliferation was defined as 1 in cells transfected with NC. Overall p is <0.0001, driven by si_A2_ni which is significantly different from NC (p<0.0001, T-test), but also significantly different from all others. No other siRNAs were significantly different from NC or MATra in the MTS experiment.
Discussion
Severe OI is a debilitating and potentially life-threatening genetic disorder with no satisfactory or curative treatment. Bisphosphonate therapy is standard care for children with moderate to severe OI and has been shown to improve both bone phenotype and increase vertebral DXA Z-scores [23,24]. However, the reduction of long bone fractures and improved strength, motor function and pain management that was initially reported in observational trials is not supported by controlled trials [7,24,25] and patients are thus still severely affected by bone fragility. There are also concerns of high cumulative doses of bisphosphonates in patients treated from infancy. There are small-scale studies and case reports on stem cell transplantation, bone marrow transplantation and gene therapy in patients with OI and in mouse models with varying results [26-30] but to date no clinically available therapy has developed out of these publications. This study was aimed at investigating a novel genetic therapeutic approach for treating or limiting the severity of this disease. Targeting common polymorphisms in contrast to individual specific mutations in collagen type I transcripts is an attractive mutation independent approach; a small number of siRNAs directed at common heterozygous alleles in COL1A1 and COL1A2 could be used for a large proportion of patients with OI, opposed to developing hundreds of siRNAs in a mutation direct approach. Nevertheless, polymorphisms are very rare in COL1A1 mRNA and there are only a few common SNPs in COL1A2 transcripts. Furthermore both genes are quite repetitive, creating difficulties designing target-specific siRNAs, which may be hard to overcome.
We have previously shown that it is possible to preferentially shift the allele ratio of COL1A2 mRNA by targeting an exonic SNP[10]. However, the best allele ratio observed for the most optimal siRNA targeting the COL1A2 SNP was merely 0.34 compared to e.g. allele ratios of 0.05 and 0.01 for the two COL1A2-indel targeting siRNAs presented here. Furthermore, in our previous study quantitative PCR results insinuated silencing also of the non-targeted allele; the SNP allele ratio of 0.34 for the middle concentration of siRNA was accompanied by an overall silencing of COL1A2 mRNA as great as 77%. In this study the allele ratios are greatly improved and total mRNA levels are closer to what is expected following truly allele discriminatory silencing, supporting the hypothesis that utilizing indels renders higher allele specificity. For the current siRNA targeting e.g the COL1A2 non-insertion allele the overall silencing was 49%, very close to the desired 50%, implying a much more stringent allele-specificity.
In this study we show that there are common indels in the 3'UTRs of both COL1A1 and COL1A2 with heterozygote genotype frequencies in the Swedish population of approximately 50%, and that these can serve as targets for allele dependent mRNA repression. As the two alleles in cells heterozygous for these indels differ by several nucleotides at the siRNA target site, marked allele preferential silencing should be possible, which was subsequently confirmed by the results presented in this study. COL1A1 allele ratios were not markedly improved for the higher 0.6µg siRNA dose compared to the lower 0.4µg dose; however, mRNA decrease past the desired 50% reduction was even more pronounced for the higher dose, suggesting that concentration is pivotal for allele specificity. At too high concentrations, spill over on the non-targeted allele is believed to render a general decrease in mRNA abundance, exemplified by the 0.6µg and 0.4µg doses of si_A1_i. The optimal siRNA concentrations and the most effective siRNA sequences still need to be determined, but these results can be viewed as proof of principle.
Our results speak in favor of utilizing siRNAs for OI-treatment as the ratio of mutated relative normal fibrils could be significantly reduced. The si_A1_i allele ratio of 0.09 (0.4µg dose) corresponds to approximately 1:12 mutated α1-chains, which would substantially reduce the number of fibrils affected by mutation compared to the untreated patient with 1:2 mutated α1-chains and consequently only 25% normal fibrils. For si_A2_ni (0.6µg dose) the allele ratio of 0.01 amounts to approximately 1:100 mutated fibrils. As the collagen type I fibrils contain only one α2-chain, this translates to 99% normal fibrils and 1% containing a mutated α2-chain compared to the untreated 50% mutated fibrils. The A2_ni siRNA would thus in theory completely cure the patient as null alleles for COL1A2 are asymptomatic [8]. In addition, the COL1A2 mRNA relative level in this experiment was the expected 51% according to the mRNA abundance formula described above (0.5+(0.5 x allele ratio)). However, as these experiments were performed on primary bone cells from healthy donors it was not possible to assess any change in collagen fiber formation in this setting. As optimal dose and incubation time have been established here, the next step would be transfection of primary bone cells from a patient with severe dominant OI, heterozygous for the appropriate indel.
Studies of mosaic carriers of OI-causing mutations [31,32] and bone marrow transplantation experiments in patients with OI and murine OI-models have shown that even a very small increase in cells producing normal collagen can have a striking effect on bone phenotype [27,33,34]. This clearly supports that the relative increase in normal collagen described above could have dramatic effects on bone health. Furthermore, it is well known that too little albeit normal collagen is favorable compared to mutated collagen[1]. Consequently, the clear decrease detected in one allele should translate into a milder phenotype in severe dominant OI.
According to the time course study the allele ratio shift was maximized around 72 hours post transfection (Figure 4) for all siRNAs, although the results are more variable for COL1A2 which is most likely due to the loss of replicates. However, all siRNAs are unique and results could differ between siRNAs and cell types. A significant effect is observed for all siRNAs at the last studied time point (216 hours), and in future studies it would be necessary to determine time-point for complete cessation of siRNA effect as well as appropriate dosage interval to maintain a desirable steady-state. Our approach of a transient conversion of a fraction of bone cells could have a significant effect on the phenotype, with the benefit of avoiding the risk of malignant transformation that could potentially be incurred by e.g. retroviruses permanently incorporating siRNA in the genome.
We established the frequency of heterozygotes for the two studied indels in cohorts of Swedish populations to be close to 50%. In addition, the heterozygote frequency for rs3840870 in COL1A1 was determined to be 46% in a published cohort of 120 unrelated controls [35] and data is also available for different ethnicities in the 1000 Genomes Project (http://www.1000genomes.org/). Allele frequencies of non-insertion vs. insertion for rs3917 in COL1A2 are also available in the National Centre for Biotechnology Information (NCBI) dbSNP-database (http://www.ncbi.nlm.nih.gov/projects/SNP/), and in the 1000 Genomes Project and heterozygous genotype is between 11% and 49% in different populations. See table 1 for all heterozygote frequencies. It can thus be assumed that the siRNAs presented in this study could be used in several other populations.
Moreover, the results presented here are of interest not only for the OI-field, but a similar approach could be employed for treating other dominant diseases where a heterozygous indel is present. Allele specific silencing has been studied utilizing several heterozygous SNPs in Huntington's disease[36-38] and a mutation direct approach has been employed in e.g. epidermolysis bullosa simplex[39], Parkinson's disease [40] and vascular Ehlers-Danlos syndrome[20]. The causative genes for these diseases could be investigated for indels, potentially useful as handles for RNAi; for instance in COL3A1 that causes vascular Ehler-Danlos syndrome the indel rs13306265 is reported at a heterozygous frequency of 0.375 in the NCBI dbSNP-database (address stated above).
Nevertheless, several obstacles remain to be overcome before truly allele specific siRNAs that render 50% silencing of COL1A1 or COL1A2 can be tested in clinical trials. SiRNA sequences will need to be optimized to achieve maximum efficiency and specificity as the base composition is of utmost importance for RNAi outcome. Furthermore, siRNAs are likely to contain sequences partially complementary to other genes, which necessitates excluding undesirable and potentially deleterious off-target effects. In order to assess effects of transfection method and viability of siRNA transfected cells an MTS assay was performed, and a slightly increased viability of si_a2_ni treated cells was observed suggesting possible off-target effects. All siRNA intended for in vivo use have to be scrutinized for off-target effects extensively, while the aim in the present study was to investigate the use of highly polymorphic small indels as targets for allele specific silencing. Partial complementarity was found for targets other than collagen type I when siRNAs were analyzed by the complementarity software MiRanda, freely available online, and as a next step hits with a high score should be investigated further e.g. by microarray.
Furthermore, an appropriate vehicle for delivery of the siRNAs in vitro and eventually in vivo will be essential and it will also be necessary to determine how to administer siRNAs specifically to the target cells in sufficient quantity. Viral vectors expressing target tissue specific short hairpin RNAs (shRNAs) as well as aptamer-shRNA chimaeras[41] may be interesting to explore in order to express siRNAs specifically in certain cell types. An especially promising approach could be the use of siRNA bound to atelocollagen [42] as this complex is resistant to nucleases and is transduced efficiently into cells, thereby allowing long-term gene silencing [43]. Atelocollagen shows neither antigenicity nor toxicity in animals, since antigenic telopeptides have been eliminated by pepsin digestion [44]. In a mouse model of metastatic prostate cancer detected by in vivo bioluminescence, mice receiving a luciferase-siRNA-atelocollagen complex, showed an 80-90% decreased bioluminescence at one day post treatment in the whole body, including the bone metastases, when compared with before treatment[45]. An additional benefit of atelocollagen is that concentration control, which is of utmost importance for allele specificity, should be feasible with this approach.
In conclusion, the results presented here show that allele specific silencing of COL1A1 and COL1A2 is possible in human bone derived cells using siRNAs targeting common indels in the 3'UTR. This is a novel approach, highly effective in vitro, and is a promising step towards personalized RNAi therapy in severe OI.
Acknowledgements
We thank Anna-Lena Johansson for skillful technical assistance.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Willing MC, Pruchno CJ, Atkinson M, Byers PH. Osteogenesis imperfecta type I is commonly due to a COL1A1 null allele of type I collagen. Am J Hum Genet. 1992;51:508-515
2. Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16:101-116
3. Sillence DO, Rimoin DL, Danks DM. Clinical variability in osteogenesis imperfecta-variable expressivity or genetic heterogeneity. Birth Defects Orig Artic Ser. 1979;15:113-129
4. Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD. et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209-221
5. Byers PH, Pyott SM. Recessively inherited forms of osteogenesis imperfecta. Annu Rev Genet. 2012;46:475-497
6. Phillini RT CA, STeiner RD. Bisphosphonate therapy for osteogenesis imperfecta. Cochrane Database Syst. Rev. 2008
7. Castillo H, Samson-Fang L. Effects of bisphosphonates in children with osteogenesis imperfecta: an AACPDM systematic review. Dev Med Child Neurol. 2009;51:17-29
8. Schwarze U, Hata R, McKusick VA, Shinkai H, Hoyme HE. et al. Rare autosomal recessive cardiac valvular form of Ehlers-Danlos syndrome results from mutations in the COL1A2 gene that activate the nonsense-mediated RNA decay pathway. Am J Hum Genet. 2004;74:917-930
9. Millington-Ward S, McMahon HP, Allen D, Tuohy G, Kiang AS. et al. RNAi of COL1A1 in mesenchymal progenitor cells. Eur J Hum Genet. 2004;12:864-866
10. Lindahl K, Rubin CJ, Kindmark A, Ljunggren O. Allele dependent silencing of COL1A2 using small interfering RNAs. Int J Med Sci. 2008;5:361-365
11. Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689-696
12. Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950-952
13. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498
14. Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D. et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111-114
15. Liao H, Irvine AD, Macewen CJ, Weed KH, Porter L. et al. Development of allele-specific therapeutic siRNA in Meesmann epithelial corneal dystrophy. PLoS One. 2011;6:e28582
16. Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J. et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140
17. Hickerson RP, Smith FJ, Reeves RE, Contag CH, Leake D. et al. Single-nucleotide-specific siRNA targeting in a dominant-negative skin model. J Invest Dermatol. 2008;128:594-605
18. Dykxhoorn DM, Schlehuber LD, London IM, Lieberman J. Determinants of specific RNA interference-mediated silencing of human beta-globin alleles differing by a single nucleotide polymorphism. Proc Natl Acad Sci U S A. 2006;103:5953-5958
19. Leslie Pedrioli DM, Fu DJ, Gonzalez-Gonzalez E, Contag CH, Kaspar RL. et al. Generic and personalized RNAi-based therapeutics for a dominant-negative epidermal fragility disorder. J Invest Dermatol. 2012;132:1627-1635
20. Muller GA, Hansen U, Xu Z, Griswold B, Talan MI. et al. Allele-specific siRNA knockdown as a personalized treatment strategy for vascular Ehlers-Danlos syndrome in human fibroblasts. FASEB J. 2012;26:668-677
21. Grundberg E, Lau EM, Pastinen T, Kindmark A, Nilsson O. et al. Vitamin D receptor 3' haplotypes are unequally expressed in primary human bone cells and associated with increased fracture risk: the MrOS Study in Sweden and Hong Kong. J Bone Miner Res. 2007;22:832-840
22. Ge B, Gurd S, Gaudin T, Dore C, Lepage P. et al. Survey of allelic expression using EST mining. Genome Res. 2005;15:1584-1591
23. Bachrach LK, Ward LM. Clinical review 1: Bisphosphonate use in childhood osteoporosis. J Clin Endocrinol Metab. 2009;94:400-409
24. Phillipi CA, Remmington T, Steiner RD. Bisphosphonate therapy for osteogenesis imperfecta. Cochrane Database Syst Rev. 2008:CD005088
25. Letocha AD, Cintas HL, Troendle JF, Reynolds JC, Cann CE. et al. Controlled trial of pamidronate in children with types III and osteogenesis IV. imperfecta confirms vertebral gains but not short-term functional improvement. J Bone Miner Res. 2005;20:977-986
26. Mehrotra M, Rosol M, Ogawa M, Larue AC. Amelioration of a mouse model of osteogenesis imperfecta with hematopoietic stem cell transplantation: microcomputed tomography studies. Exp Hematol. 2010;38:593-602
27. Panaroni C, Gioia R, Lupi A, Besio R, Goldstein SA. et al. In utero transplantation of adult bone marrow decreases perinatal lethality and rescues the bone phenotype in the knockin murine model for classical, dominant osteogenesis imperfecta. Blood. 2009;114:459-468
28. Li F, Wang X, Niyibizi C. Distribution of single-cell expanded marrow derived progenitors in a developing mouse model of osteogenesis imperfecta following systemic transplantation. Stem Cells. 2007;25:3183-3193
29. Guillot PV, Abass O, Bassett JH, Shefelbine SJ, Bou-Gharios G. et al. Intrauterine transplantation of human fetal mesenchymal stem cells from first-trimester blood repairs bone and reduces fractures in osteogenesis imperfecta mice. Blood. 2008;111:1717-1725
30. Le Blanc K, Gotherstrom C, Ringden O, Hassan M, McMahon R. et al. Fetal mesenchymal stem-cell engraftment in bone after in utero transplantation in a patient with severe osteogenesis imperfecta. Transplantation. 2005;79:1607-1614
31. Raghunath M, Mackay K, Dalgleish R, Steinmann B. Genetic counselling on brittle grounds: recurring osteogenesis imperfecta due to parental mosaicism for a dominant mutation. Eur J Pediatr. 1995;154:123-129
32. Cabral WA, Marini JC. High proportion of mutant osteoblasts is compatible with normal skeletal function in mosaic carriers of osteogenesis imperfecta. Am J Hum Genet. 2004;74:752-760
33. Horwitz EM, Prockop DJ, Gordon PL, Koo WW, Fitzpatrick LA. et al. Clinical responses to bone marrow transplantation in children with severe osteogenesis imperfecta. Blood. 2001;97:1227-1231
34. Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD. et al. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci U S A. 2002;99:8932-8937
35. Nuytinck L, Coppin C, De Paepe A. A four base pair insertion polymorphism in the 3' untranslated region of the COL1A1 gene is highly informative for null-allele testing in patients with osteogenesis imperfecta type I. Matrix Biol. 1998;16:349-352
36. Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F. et al. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217:312-319
37. Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S. et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178-2185
38. Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W. et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol. 2009;19:774-778
39. Atkinson SD, McGilligan VE, Liao H, Szeverenyi I, Smith FJ. et al. Development of allele-specific therapeutic siRNA for keratin 5 mutations in epidermolysis bullosa simplex. J Invest Dermatol. 2011;131:2079-2086
40. de Ynigo-Mojado L, Martin-Ruiz I, Sutherland JD. Efficient allele-specific targeting of LRRK2 R1441 mutations mediated by RNAi. PLoS One. 2011;6:e21352
41. McNamara JO 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE. et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005-1015
42. Ashihara E, Kawata E, Maekawa T. Future prospect of RNA interference for cancer therapies. Curr Drug Targets. 2010;11:345-360
43. Minakuchi Y, Takeshita F, Kosaka N, Sasaki H, Yamamoto Y. et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32:e109
44. Ochiya T, Takahama Y, Nagahara S, Sumita Y, Hisada A. et al. New delivery system for plasmid DNA in vivo using atelocollagen as a carrier material: the Minipellet. Nat Med. 1999;5:707-710
45. Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H. et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci U S A. 2005;102:12177-12182
Footnotes
Rs3840870 and rs35521741 refer to the same polymorphism. This COL1A1 insertion will be denoted rs3840870 in this paper.
Author contact
![]() Corresponding author: Dr. Katarina Lindahl; katarina.lindahluu.se; MMS, ing. 40, Akademiska sjukhuset, 751 85 Uppsala, Sweden. Phone: (+46)707487875
Corresponding author: Dr. Katarina Lindahl; katarina.lindahluu.se; MMS, ing. 40, Akademiska sjukhuset, 751 85 Uppsala, Sweden. Phone: (+46)707487875

 Global reach, higher impact
Global reach, higher impact