Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(9):1181-1192. doi:10.7150/ijms.6657 This issue Cite
Research Paper
CXCL12/CXCR4 Signal Axis Plays an Important Role in Mediating Bone Morphogenetic Protein 9-induced Osteogenic Differentiation of Mesenchymal Stem Cells
1. Key Laboratory of Laboratory Medical Diagnostics, Ministry of Education, Department of Laboratory Medicine, Chongqing Medical University, Chongqing 400016, China;
2. Department of Laboratory Medicine, Jinan Sixth Hospital, Zhangqiu250200, China.
* These authors contributed equally to this work.
Received 2013-5-9; Accepted 2013-6-11; Published 2013-7-17
Abstract
Mesenchymal progenitor stem cells (MPCs) are a group of bone marrow stromal progenitor cells processing osteogenic, chondrogenic, adipogenic and myogenic lineages differentiations. Previous studies have demonstrated that bone morphogeneic protein 9(BMP9) is one of the most osteogenic BMPs both in vitro and in vivo, however, the underlying molecular mechanism of osteogenesis induced by BMP9 is needed to be deep explored. Here, we used the recombinant adenoviruses assay to introduce BMP9 into C3H10T1/2 mesenchymal stem cells to elucidate the role of CXCL12/CXCR4 signal axis during BMP9-incuced osteogenic differentiation. The results showed that CXCL12 and CXCR4 expressions were down-regulated at the stage of BMP9-induced osteogenic differentiation, in a dose- and time-dependent. Pretreatment of C3H10T1/2 cells with CXCL12/CXCR4 could significantly affect the early and mid osteogenic markers alkaline phosphatase (ALP), osteocalcin (OCN), the transcription factors of Runx2, Osx, Plzf and Dlx5 expression, through activating the Smad, MAPK signaling pathway. Addition of exogenous CXCL12 did not affect the changes of the late osteogenic marker calcium deposition. Thus, our findings suggest a co-requirement of the CXCL12/CXCR4 signal axis in BMP9-induced the early- and mid-process of osteogenic differentiation of MSCs.
Keywords: CXCL 12, CXCR4, Bone Morphogenetic Protein 9, Mesenchymal Stem Cells, Osteogenic Differentiation.
1. Introduction
Mesenchymal progenitor cells (MPCs) are non-hematopoietic and multipotent cells [1] that have the capacity to self-new [2] and differentiate into diverse cell lineages including myoblasts, chondrocytes, adipocytes and osteoblasts [3, 4]. Although MPCs have been found in various tissues, including brain, liver, gut, skeletal muscle, and adipose [5, 6], one of the major sources is bone marrow, representing 0.001-0.01% of the nucleated cells [7]. While MPCs isolating from various tissues share many similar characteristics, they exhibit minor differences in their expression profile and differentiation potential [6]. Osteogenesis is a sequential cascade that recapitulates the most, if not all, of the cellular events occurring during embryonic and skeletal development [8]. Osteogenic differentiation of MPCs is a complex, tightly regulated progress that critical for proper bone formation and is defined by several endogenous and environmental factors, such as BMPs, Runx2, PPARγ, and Wnt/β-catenin [9].
Bone morphogenetic proteins (BMPs), members of transforming growth factors β (TGFβ) superfamily, are known to play pivotal roles in regulating cell proliferation and differentiation during development [10]. To date, at least 20 members of BMPs have been identified [11]. BMPs fulfill their signaling activity by interacting with two trans-membrane serine/threonine kinase receptors, BMPR type I and II [12]. These activated receptor kinase transduce the BMP signaling by phosphorylating the transcription factor Smad1/5/8, which in turn form a heterodimeric complex with Smad4 in the nucleus and regulate the transcription of target genes [13]. Besides Smads, the mitogen activated protein kinases (MAPKs) have been demonstrated to be involved in transmitting intracellular signaling of BMPs, including the extracellular signal-regulated kinases 1/2 (Erk1/2), Erk5, the Jun amino-terminal kinases (JNKs), and the p38 MAPKs[14]. Several forms of recombinant BMPs (rhBMPs), the most notably rhBMP2 and rhBMP7, have been used as adjunctive therapy in clinical [15]. Recent studies have indicated that BMP9, one of the least studied BMPs, showing the most potent osteogenic activity in MPCs [12, 16]. Despite these valuable discoveries, the molecular mechanism of BMP9-induced osteogenic differentiation is still largely obscure and warrants extensive studies.
The chemokine CXCL12 (stromal derived factors 1, SDF-1) belong to the C-X-C chemokine family and was originally isolated from a murine bone marrow stromal cell line as a growth-stimulating factor for the B cell precursor clone [17]. CXCL12 is widely and constitutively expressed by numerous tissues, juxtaposed in its expression with that of its cognate receptors, CXCR4 and CXCR7 [18]. Interactions between CXCL12 and CXCR4 have been shown to modulate the trafficking and proper engraftment of hematopoietic stem cell (HSCs) and reconstitution of hematopoiesis [19]. Other studies have implicated CXCL12 as an important factor in promoting the survival and migration of circulating tissue-specific progenitors identified for muscle, neural, and liver, implicating CXCL12 as an important maintenance factors in postnatal tissue repair [20]. It has also been proved that CXCR4 is expressed in malignant tumors and the CXCL12/CXCR4 interaction is involved in tumor progression particularly brain, breast tumors and osteosarcoma [21]. Recent studies demonstrated a regulatory role of CXCL12 in osteogenic differentiation induced by BMP2 in several sources of MPCs, including C2C12, ST2 [22] and bone-marrow derived mesenchymal stromal cells [23]. However, it remains unknown whether CXCL12/CXCR4 signaling axis is involved in the osteogenic differentiation induced by BMP9.
Based on these premises, we further hypothesized that CXCL12/CXCR4 signaling axis is required for BMP9-induced o osteogenic differentiation of MPCs. We find that BMP9-induced early osteogenic markers, such as alkaline phosphatase (ALP), and middle osteogenic markers, such as osteocaclin (OCN), are synergistically enhanced by CXCL12/CXCR4 signaling axis in vitro, instead of the late osteogenic marker, calcium deposition. And the regulatory effect of CXCL12/CXCR4 signaling axis is mediated via intracellular Smad and MAPK activation. These results strongly suggest that CXCL12/CXCR4 signaling axis can effectively augment BMP9-induced osteogenic differentiation of MPCs.
2. Materials and Methods
2.1 Cell line and culture conditions
HCT116, HEK293, HS5 and C3H10T1/2 mesenchymal stem cells were purchased from American Type Culture Collection and maintained in complete Dulbeccos modified Eagle medium (DMEM, Hyclone, USA) supplemented with 10% fetal bovine serum (FBS, GIBCO, USA) and 100 units/ml penicillin/streptomycin in a 5% CO2-humidified incubator at 37 °C.
2.2 Recombinant adenoviruses over expressing RFP, GFP, BMP9, CXCL12 and suppressing CXCR4
Recombinant adenoviruses were generated using AdEasy system as demonstrated. Briefly, the coding regions of human BMP9 and human CXCL12 were amplified by PCR and subcloned into an adenoviral shuttle vector pAdTrace-TOX and subsequently used to generate recombinant adenoviruses in HEK293 cells. The small interfering RNA (siRNA) target sites against the human CXCR4 coding region were cloned into the homemade siRNA expression pSES adenoviral shuttle vector to generate recombinant adenovirus in HEK293 cells. The resulting adenoviruses were packaged and designated as AdBMP9, Ad-CXCL12and Adsi-CXCR4. Ad-BMP9 expressed green fluorescent protein (GFP), whereas Ad-CXCL12 and Adsi-CXCR4 express red fluorescent protein (RFP) as a marker for monitoring infection efficiency. Analogous adenovirus expressing only monomeric RFP (Ad-RFP) or GFP (Ad-GFP) were used as controls.
2.3 Preparation of conditioned medium
BMP9 conditioned medium were prepared as described. Briefly, subconfluent HCT116 cells in 75-cm2 flasks were infected with an optimal titer of Ad-BMP9. At 15 hours after infection, the culture medium was changed to serum-free DMEM. Conditioned medium was collected at 48 hours after infection and used immediately.
2.4 Total RNA isolation, RT-PCR, and real time quantitative PCR (qPCR) analysis
Subconfluent cells were seeded in 75-cm2 cell culture flasks in a medium supplemented with 0.5% FBS with or without adenoviruses infection. Total RNA was isolated using TRIZOL Reagents (Invitrogen, USA) according to the manufacturer's instructions. RNA quality and integrity were verified using the NanoDrop100 UV-Vis spectrophotometer (Thermo Scientific, USA). 2mg of total RNA were used to generate cDNA templates by reverse transcription with hexamer primers and the Prime Script kit (TaKaRa, Japan). The first strand cDNA products were further diluted and used as qPCR templates. SYBR Green I dye-based qPCR analysis was carried out using the Rotor-Gene 6000 Real-Time PCR machine (Corbett Research, Australia). Primers used are listed in Table 1. Standard curves were used to determine RNA levels, and values were normalized to GAPDH. Triplicate reactions were carried out for each sample.
2.5 Western blotting analysis
Western blotting experiments were carried out essentially as previously described. Briefly, cells first washed with ice-cold PBS and lysed in 200μl RIPA lysis buffer (BiYunTian, China). Cell lysates were boiled for 10 min. The total cellular protein concentration of each supernatant was determined by using the NanoDrop100 UV-Vis Spectrophotometer (Thermo Scientific, USA) and loaded onto a 10% gradient SDS-PAGE. After electrophoresis separation, proteins were transferred to PVDF membranes (Millipore, USA). Membrane was blocked with Super-Block Blocking Buffer, and probed with the primary antibody, followed by incubation with a secondary antibody (ZhongShan Goldenbridge Biotechnology, China) conjugated with horseradish peroxidase. The proteins of interest were visualized using space chemoluminescence reagent (Pierce, USA) in the ChemiDoc™ XRS+ system (Bio-Rad, USA). Primary antibodies were obtained from Santa Cruz, as follows: Anti-CXCR4, anti-phosphor-Smad1/5/8, anti-Smad1/5/8, anti-phosphor-ERK1/2, anti-ERK1/2 and anti-β-actin.
2.6 ELISA
The protein concentrations of CXCL12 in the cell lysates was determined by using ELISA Kits (Raybio, China). Briefly, samples were added to the coated wells with CXCL12 mAb of 96-well plates and incubated for 2h at room temperature. They were washed three times and incubated with an HRP-linked streptavidin solution for 30 minutes at 37℃. The absorbance was measured at 450nm on the microplate reader (Sunrise Remote, Switzerland) immediately. All the experiments were repeated for three times.
2.7 Alkaline Phosphatase (ALP) Assay
The cells in the transfection groups and non-transfection groups were harvested. ALP activity was assessed at day 7 after transfection by a modified Great Escape SEAP Chemiluminescence assay (BD Clontech, CA) and/or histochemical staining assay (using a mixture of 0.1 mg/ml napthol AS-MX phosphate and 0.6 mg/ml Fast Blue BB salt) as described. For the chemiluminescence assays, each assay condition was performed in triplicate and the results were repeated in for at least three times. ALP activity was normalized by total cellular protein concentrations among the samples. ALP staining assays were observed under light microscope.
The sequence of PCR primers for each gene.

2.8 Immunohistochemical staining
Immunohistochemical staining procedure was constructed as described. Briefly, cells were seeded in 24-well cell culture plates. At day 14 after cultured, cells were fixed in 4% paraformaldehyde for 10mins, then washed twice with PBS and penetrated with 0.3% TritonX-100 (prepared in PBS) at room temperature for 10mins, followed by incubation with an anti-OCN antibody (Santa Cruz, USA), and then with biotin-labeled secondary antibody (Zhongshan Goldenbridge Biotech, China) at room temperature for 30mins. Finally, cells were stained with DAB for 3-5mins and then washed with ddH2O briefly. In the negative control, the primary antibody was replaced with PBS. Cells with brown granules in the cytoplasm were regarded to be positive for target protein. Each assay condition was performed in triplicate and the results were repeated in for least three times. Observe and take images under microscope.
2.9 Alizarin Red S staining
Calcium deposition was detected by Alizarin Red S staining, as described previously. Briefly, cells were seeded in 24-well cell culture plates and cultured in the presence of ascorbic acid (50μg/ml) and β-glycerophosphate (10mmol/l). At day 21 after cultured, cells were fixed with 0.05% (v/v) glutaraldehyde at room temperature for 10 min. After being washed with distilled water, fixed cells were incubated with 0.4% Alizarin Red S (Sigma-Aldrich, USA) for 5 min, followed by extensive washing with distilled water. Each assay condition was performed in triplicate and the results were repeated in for least three times. The staining of calcium mineral deposits was recorded under bright field microscopy.
2.10 Transfection and Luciferase Reporter Assay
Luciferase reporter assay was carried out as described previously. In brief, the exponentially growing cells were seeded in 25-cm2 culture flasks and transfected with 3.0μg per flask of BMP receptor Smad-responsive luciferase reporter, p12xSBE-Luc reporter (kindly provided by Dr. Di Chen of University of Rochester Medical Center), using LipofectamineTM 2000 (Invitrogen, USA). At 16h post transfection, cells were replated in 6-well cell culture plates and treated with processing factors or solvent control. At 36h after treatment, cells were lysed and subjected to luciferase activity assays using Luciferase Assay Kit (Promega, USA). Each assay condition was done in triplicate and the results were repeated in for least three times. Luciferase activity was normalized by total cellular protein concentrations among the samples.
2.11 Statistical analysis
Analysis of mRNA relative expression was performed using real-time quantitative PCR and 2-∆∆CT method. Microsoft Excel was used to calculate standard deviations (SD) and statistically significant differences between samples using the independent sample t-test. For all quantitative assays, each assay condition was performed in triplicate and the results were repeated in at least three independent experiments. All collected data were subjected to statistical analysis. A P-value<0.05 was defined as statistically significance. All data were analyzed by using SPSS 15.0.
3. Result
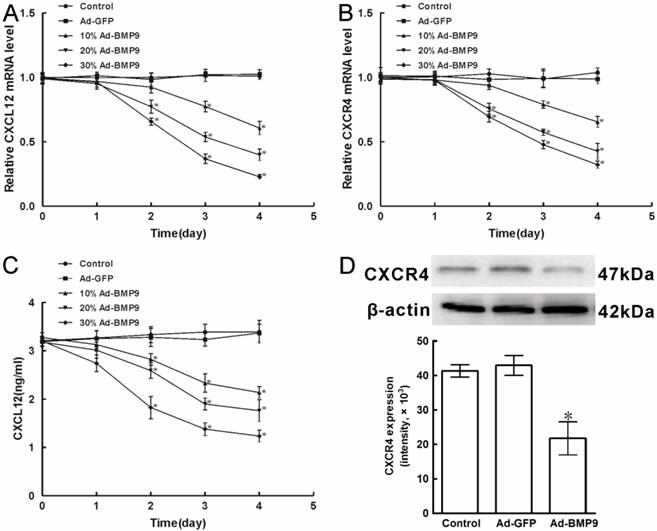
3.1 Expressing of CXCL12 and CXCR4 decreased during BMP9-induced osteogenic differentiation in C3H10T1/2 cells
Previously, it is reported that C3H10T1/2 cells express CXCL12 mRNA and protein. However, the regulation and potential role of CXCL12 and CXCR4 in BMP9-induced osteogenic differentiation of C3H10T1/2 cells remained poorly understood. Then we analyzed the expression of CXCL12 and CXCR4 by real time PCR. Different titers of exogenous BMP9 were introduced into C3H10T1/2 cells using recombinant adenovirus assay as the 24h fluorescent infection efficiency 10%, 20%, and 30%, respectively. We found that when Ad-BMP9 was introduced into C3H10T1/2 cells for 1 day, no significant differences in the mRNA levels of CXCL12 and CXCR4 could be observed compared with base-line levels in control cells stimulated with Ad-GFP. And stimulation with 20%Ad-BMP9 and 30%Ad-BMP9 for 2 days reduced the mRNA levels of CXCL12 by 22.9% and 34.4%, whereas the mRNA levels of CXCR4 was reduced by 24.4% and 30.7%, respectively. And stimulation with 10%Ad-BMP9, 20%Ad-BMP9 and 30%Ad-BMP9 for 4 days reduced the mRNA levels of CXCL12 by 39.4%, 60.2% and 77.2%, whereas the mRNA levels of CXCR4 was reduced by 34.6%, 57.4% and 67.9%, respectively (p<0.01, Figure 1A, B).
At the same time, the protein levels of CXCL12 and CXCR4 were analyzed by ELISA and Western Blot, respectively. When compared with the control cells introduced with Ad-GFP, stimulation with different titers of Ad-BMP9 for 1 day did not significantly alter the level of CXCL12 protein, but CXCL12 synthesis declined 33.4%, 45.1%, and 61.5% upon 10%Ad-BMP9, 20%Ad-BMP9, and 30%Ad-BMP9 stimulation for 4 days, respectively (p<0.01, Figure 1C). Furthermore, Western Blot results showed that 30%Ad-BMP9 stimulation for 4 days decreased the level of CXCR4 protein by 47.3%, relative to control cells (p<0.01, Figure 1D). These findings suggested that CXCL12 and CXCR4 were both expressed in mRNA and protein levels of C3H10T1/2 cells, while the expressing was able to be reduced by the stimulation of BMP9, in a time- and dose-dependent.
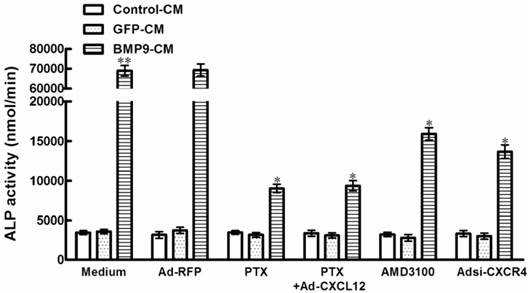
3.2 Blocking the CXCR4 expressing decreased the ALP activity induced by BMP9 in C3H10T1/2 cells
In order to determine whether CXCL12/CXCR4 signal axis is directly involved in the osteogenic differentiation of C3H10T1/2 cells, we tested how inhibition of CXCR4 expression affects BMP9-induced ALP activity, a well-established marker of early osteogenic differentiation. It is well established that chemokines activate directional cell movement via Gi protein-coupled signal transduction pathways. In the case of the chemokine CXCL12, this signal is mediated through its sole GPCR on the cell surface, CXCR4. Pertussis toxin (PTX) is a broad spectrum inhibitor that uncouples Gi proteins from GPCRs. Real time PCR results suggested that treatment with 200ng/mL PTX was able to led a significant 86.9% inhibition of ALP activity in C3H10T1/2 cells induced by BMP9-CM, when compared with the cells treated by BMP9-CM alone(p<0.01, Figure 2). In addition, the suppressed effect of PTX on BMP9-induced ALP activity was not able to be rescued by the stimulation of exogenous CXCL12 using recombinant adenovirus assay (Figure 2).
To provide additional evidence for the role of CXCR4 in BMP9-induced osteogenic differentiation, we studied that the CXCR4 antagonist, AMD3100, reduce BMP9-induced ALP activity by 76.9% in C3H10T1/2 cells compared with the cells treated by GFP-CM. And in the presence of Adsi-CXCR4, BMP9-induced ALP activity was suppressed by 80.3% (p<0.01, Figure 2). Whereas there were no significant differences in ALP activity levels in cells treated with PTX, AMD3100, and Adsi-CXCR4 alone when compared with control (Figure 2). These data indicated that during the procedure of BMP-induced osteogenic differentiation, CXCR4 is essential.
The expression of CXCL12 and CXCR4 affected by BMP9 in C3H10T1/2 cells. As controls, cells were maintained in culture medium without adenoviruses infection or stimulated with Ad-GFP. A, B, The level of CXCL12 and CXCR4 mRNA expression affected by Ad-BMP9 were both detected by qPCR and normalized to GAPDH. C, The level of CXCL12 protein expression was evaluated by ELISA. D, The level of CXCR4 protein expression in C3H10T1/2 cells affected by 30%Ad-BMP9 for 4 days was detected by Western blot. The average signal intensity was standardized to β-actin. Data were shown as means ± SD of triplicate experiments. *, p<0.01 versus the Ad-GFP group.
Blocking the expression of CXCR4 affected the BMP9-induced ALP activity in C3H10T1/2 cells. The cells were pretreated with PTX at 200ng/ml for 4h alone or after the addition of Ad-CXCL12 for 6h prior to the stimulation of BMP9-CM or GFP-CM. In addition, cells were pretreated with 400μM AMD3100 for 4h or Adsi-CXCR4 for 6h before stimulate with BMP9-CM or GFP-CM. As controls, cells were maintained in medium or stimulated with Ad-RFP. The ALP activity in C3H10T1/2 cells was detected at 7 days after the BMP9 stimulation. Data were shown as means ± SD of triplicate experiments. **and*, p<0.01 versus the Medium/GFP-CM group and Medium/BMP9-CM group, respectively.
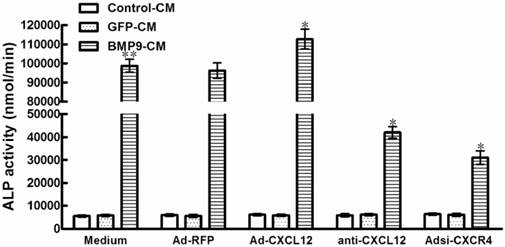
Perturbing the CXCL12/CXCR4 signal axis after the stimulation of BMP9 did not affect the ALP activity in C3H10T1/2 cells. The cells were pretreated with BMP9-CM or GFP-CM for 24h prior to the stimulation of Ad-CXCL12, anti-CXCL12 neutralizing antibody at 100μg/ml and Adsi-CXCR4, or as controls, maintained in culture medium only or stimulated with Ad-RFP. The ALP activity in C3H10T1/2 cells was detected at 7 days after the CXCL12/CXCR4 stimulation. Data were shown as means ± SD of triplicate experiments. **, p<0.01 versus the Medium/GFP-CM group.
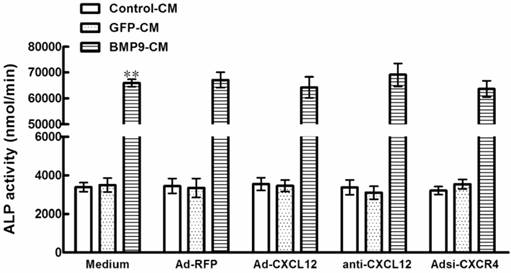
3.3 The CXCL12/CXCR4 signal axis is a prerequisite for BMP9-induced ALP activity in C3H10T1/2 cells
In the experiments described above, the CXCL12/CXCR4 signal axis of cells was perturbed before the stimulation of BMP9-CM. To confirm the involvement of CXCL12/CXCR4 signaling in BMP9-induced osteogenic differentiation in C3H10T1/2 cells, we added the BMP9-CM before the addition of Ad-CXCL12, anti-CXCL12 neutralizing antibody, and Adsi-CXCR4. No significant effects on the BMP9-dependent induction of ALP activity was able to be observed. In addition, treatment with Ad-CXCL12, anti-CXCL12 neutralizing antibody, and Adsi-CXCR4 alone did not affect ALP activity of cells when compared with control cells maintained in medium only (Figure 3). The data suggested that the CXCL12/CXCR4 signaling is a prerequisite for BMP9-induced osteogenic differentiation in C3H10T1/2 cells.
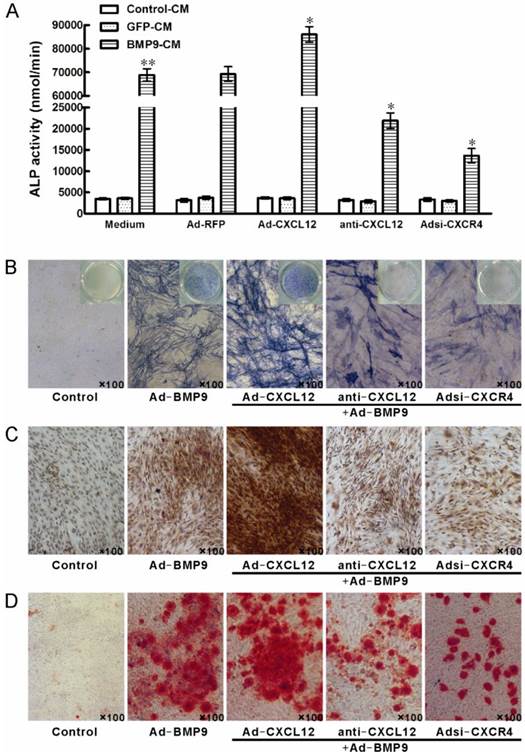
3.4 Perturbing the CXCL12/CXCR4 signal axis affected BMP9-induced osteogenic differentiation in C3H10T1/2 cells
To further document the detail roles of CXCL12 signaling in BMP9-induced osteogenic differentiation, we further analyzed whether perturbing the CXCL12/CXCR4 signal axis affected BMP9-induced osteogenic differentiation of C3H10T1/2 cells. The major data we measured were ALP quantitative assay and staining assay, OCN synthesis and calcium deposition. Our data showed that stimulation of BMP9-CM for 7 days significantly increased ALP activities by 19 folds over cells treated with GFP-CM. Adding exogenous CXCL12 via recombinant adenovirus assay prior to BMP9 stimulation further enhanced the BMP9-induced ALP activity by 24.20% compared with cells treated with BMP9-CM alone. Moreover, pretreatment with anti-CXCL12 neutralizing antibody and Adsi-CXCR4 suppressed the BMP9-induced ALP activity by 68.37% and 80.28%, respectively, when compared with cells stimulated with BMP-CM alone (p<0.01, Figure 4A). Similar results were demonstrated by the ALP staining assay (Figure 4B).
By immunocytochemistry, we also detected that prior perturbing the CXCL12/CXCR4 signal axis by anti-CXCL12 antibody or Adsi-CXCR4 for 14 days decreased the BMP9-induced OCN synthesis, and the OCN synthesis induced by BMP9 was enhanced by the pretreatment of Ad-CXCL12, respectively, when compared to cells treated with BMP9-CM alone (Figure 4C).
Perturbing the CXCL12/CXCR4 signal axis affected BMP9-induced ALP activity, OCN synthesis and calcium deposition in C3H10T1/2 cells. The cells were stimulated with Ad-CXCL12, anti-CXCL12 neutralizing antibody at 100μg/ml and Adsi-CXCR4 for 6h prior to the stimulation of BMP-CM or GFP-CM, as controls, maintained in culture medium only or stimulated with Ad-RFP. A: The ALP activity in C3H10T1/2 cells was detected at 7 days after the BMP9-CM stimulation. Data were shown as means ± SD. **and*, p<0.01 versus the Medium/GFP-CM group and Medium/BMP9-CM group, respectively. B: The ALP staining assay in C3H10T1/2 cells was detected at 7 days after the BMP9-CM stimulation (100×). C: The OCN synthesis was detected by immunocytochemistry at 14 days after the BMP9-CM stimulation (100×). D: The calcium deposition was detected by Alizarin Red staining at 21 days after the BMP9-CM stimulation (100×). Experiments were carried out in triplicate and representative results are shown.
Perturbing the CXCL12/CXCR4 signal axis affected BMP9-induced ALP activity in HS5 cells. The cells were stimulated with Ad-CXCL12, anti-CXCL12 neutralizing antibody at 100μg/ml and Adsi-CXCR4 for 6h prior to the stimulation of BMP-CM or GFP-CM, as controls, maintained in culture medium only or stimulated with Ad-RFP. The ALP activity in HS5 cells was detected at 7 days after the BMP9-CM stimulation. Data were shown as means ± SD of triplicate experiments. **and*, p<0.01 versus the Medium/GFP-CM group and Medium/BMP9-CM group, respectively.
After 21 days of BMP9 stimulation, we detected the synthesis of calcium by Alizarin Red staining. BMP9-CM stimulation increased the calcium-staining strongly. Moreover, the BMP9-stimulated calcium-staining was reduced by the pretreatment of both anti-CXCL12 neutralizing antibody and Adsi-CXCR4. However, pretreatment of exogenous Ad-CXCL12 did not enhance the level of calcium deposition induced by BMP9 compared with the cells treated with BMP9-CM alone (Figure 4D).
Together, these results intensively suggested that CXCL12/CXCR4 signal axis was involved in regulation of BMP9-induced early and middle osteogenic differentiation in C3H10T1/2 cells.
3.5 Perturbing the CXCL12/CXCR4 signal axis also affected the BMP9-induced ALP activity in HS5 cells
According to the results we got from the C3H10T1/2 cells above, we tested whether CXCL12 signaling involved in the BMP9-induced osteogenic differentiation of another cell line. In previous studies, we found that HS5 cells, which is a human bone marrow-derived stromal cell line, undergoing the function of osteogenic differentiation induced by BMP9. Our data showed that stimulation of BMP9-CM for 7 days significantly increased ALP activity over cells treated with GFP-CM. Pretreatment of HS5 cells with anti-CXCL12 neutralizing antibody and Adsi-CXCR4 reduced the BMP9 induced ALP activity by 57.47% and 68.63%, respectively, while the addition of exogenous Ad-CXCL12 prior to BMP9 stimulation enhanced the BMP9 induced ALP-activity by 14.13% when compared with the cells treated with BMP9-CM alone (p<0.01, Figure 5). The data suggested that the CXCL12/CXCR4 signal axis also played an important role in BMP9-induced osteogenic differentiation in HS5 cells compared with C3H10T1/2 cells.
3.6 Blocking the CXCL12/CXCR4 signal axis inhibited the expression of osteoblast-associated transcription factors
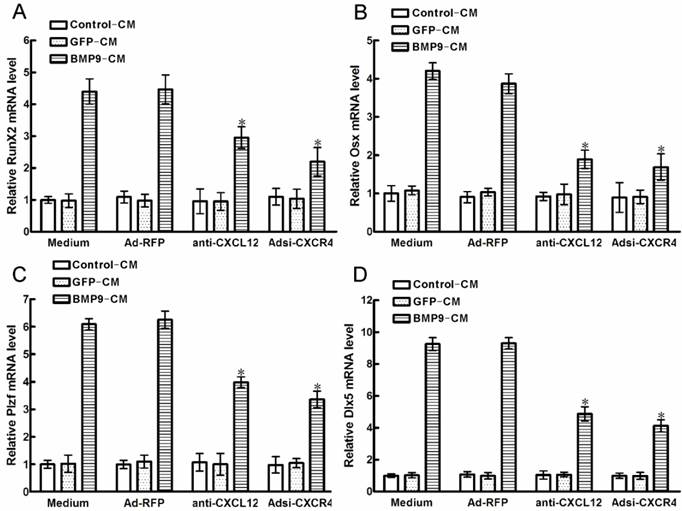
Downstream of BMP9-induced osteogenic differentiation pathways is a cascade of crucial transcription factors, such as RUNX2 and OSX, which promoted the regulation of osteogenic differentiation markers, ALP, OCN and calcium deposition, et al. Plzf is one of the early transcription factors upstream of RUNX2,while Dlx5 is one of the early transcription factors upstream of OSX. In order to determine the endoneclear mechanism of CXCL12 involved in the regulation of BMP9-induced osteogenic differentiation. We analyzed whether blocking CXCL12/CXCR4 signal axis affects the endoneclear transcription factors above.
After 6h of BMP9-CM stimulation, the time of peak expression of RUNX2 and OSX in C2C12 cells [22], we detected that all of the transcription factors were strongly increased. Pretreatment with anti-CXCL12 neutralizing antibody and Adsi-CXCR4 for 6h, strongly reduced the BMP9-induced RUNX2 expression by 33.87% and 50.77% (p<0.01, Figure 6A), and the BMP9-induced OSX expression was decreased by 51.02% and 56.32% when compared with the control cells (p<0.01, Figure 6B), respectively. At the same time, we got same tendency from the BMP9-induced Plzf and Dlx5 expression in C3H10T1/2 cells. Similarly, pretreatment with anti-CXCL12 neutralizing antibody and Adsi-CXCR4 inhibited the BMP9-induced Plzf expression by 36.31% and 46.27% (p<0.01, Figure 6C), and the Dlx5 expression was reduced by 47.64% and 55.67% (p<0.01, Figure 6D) when compared with the control cells, respectively. Collectively, these data showed that blocking the CXCL12/CXCR4 signaling inhibited the expression of BMP9-associated transcription factors, such as RUNX2, Plzf, OSX and Dlx5.
3.7 The CXCL12/CXCR4 signal axis was involved in the phosphorylation of Smad1/5/8 and ERK1/2 in C3H10T1/2 cells
A recent study by XU et al [24] demonstrated that BMP9 simultaneously promotes the activation of Smad1/5/8 and ERK1/2 in C3H10T1/2 cells. In order to further understand the molecule mechanisms of hoe CXCL12/CXCR4 signal axis affected on the BMP9-induced osteogenic differentiation, we examined how perturbing CXCL12 signaling affects the activation of Smad1/5/8 and ERK1/2 induced by BMP9.
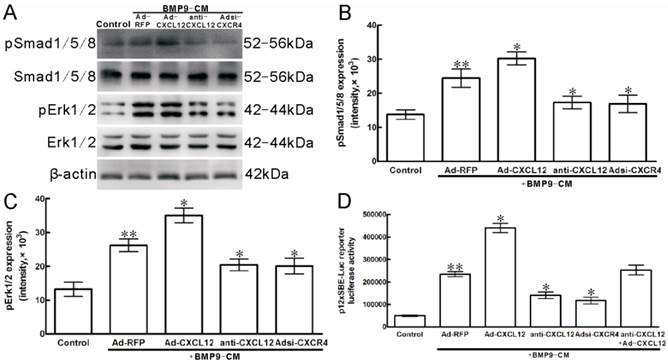
In C3H10T1/2 cells, Western Blot results showed that stimulation with BMP9-CM for 30min increased the phosphorylation of Smad1/5/8 by 2 folds when compared with the cells treated with GFP-CM (p<0.01, Figure 7A, B). Pretreatment with anti-CXCL12 neutralizing antibody and Adsi-CXCR4 decreased the BMP9-stimulated phosphorylation of Smad1/5/8 by 29.13% and 30.93% (p<0.05, Figure 7A, B), respectively, without affecting the total amounts of these transcription factors. At the same time, the addition of Ad-CXCL12 increased the BMP9-induced phosphorylation of Smad1/5/8 by 23.92% (p<0.05, Figure 7A, B).
To further ascertain that CXCL12 signaling affect the Smad1/5/8, we used the BMP Smad-responsive luciferase reporter. C3H10T1/2 cells were transfected with 12×SBE-Luc reporters and treated with anti-CXCL12 neutralizing antibody and Adsi-CXCR4 for 24h, and then stimulated with BMP9-CM. Luciferase activity was measured at 24h after BMP9-CM stimulation. The luciferase activity of treatment with BMP9-CM alone was 5 folds than that in control, respectively. The addition of anti-CXCL12 neutralizing antibody and Adsi-CXCR4 resulted in the decrease of the level of BMP9-induced luciferase by 40.01% and 50.02% (p<0.01, Figure 7D), and the anti-CXCL12 neutralizing antibody blocked was rescued by the addition of CXCL12-CM (Figure 7D), while Ad-RFP exhibited no significant effects. Moreover, the luciferase activity induced by BMP9-CM was strongly enhanced by the pretreatment of CXCL12-CM by 87.25% (p<0.01, Figure 7D).
Blocking the CXCL12/CXCR4 signal axis inhibited BMP9-induced expressions of transcription factors. The cells were stimulated with Ad-CXCL12, anti-CXCL12 neutralizing antibody at 100μg/ml and Adsi-CXCR4 for 6h prior to the stimulation of BMP-CM or GFP-CM, as controls, maintained in culture medium only or stimulated with Ad-RFP. 1day later, the cells were collected for total RNA isolation. Each assay condition was carried out in triplicate. Data were shown as means ± SD of triplicate experiments. *, p<0.01 versus the Medium/BMP9-CM group.
Perturbing the CXCL12/CXCR4 signal axis affected BMP9-dependent intracellular Smad and Erk phosphorylation in C3H10T1/2 cells. The cells were stimulated with Ad-CXCL12, anti-CXCL12 neutralizing antibody at 100μg/ml and Adsi-CXCR4 for 6h prior to the stimulation of BMP-CM for 30min, as controls, stimulated with Ad-RFP. A: Total Smad1/5/8 and Erk1/2, the phosphorylated Smad1/5/8 and Erk1/2, and β-actin were detected in cell lysates by Western blot using appropriate antibodies. B, C: Levels of pSmad1/5/8 and Erk1/2 were measured by Quantity One image software. D: The p12×SBE-luc reporter luciferase activity of C3H10T1/2 cells was measured. Data were shown as means ± SD of triplicate experiments. **and*, p<0.05 versus the control group and Ad-RFP/BMP9-CM group, respectively.
Then, we studied whether the phosphorylation of one component of the MAPK signaling pathway, ERK1/2 was involved in the procedure that CXCL12 signaling affected on the BMP9-induced osteogenic differentiation.
Western blot analysis showed that BMP9-CM treatment to the C3H10T1/2 cells for 30min strongly increased the phosphorylation of Erk1/2 by 2 folds when compared with the control cells (p<0.01, Figure 7A, C). Pretreatment with anti-CXCL12 neutralizing antibody reduced the BMP9-dependent activation of Erk1/2 by 22.01%, whereas it is reduced by 23.49% when the cells were transfected with Adsi-CXCR4 compared with the cells treated with BMP9-CM alone (p<0.05, Figure 7A, C). Similar levels of total ERK1/2 and β-action were detected in C3H10T1/2 cells.
4. Discussion
To date, BMP9 remains as one of the least characterized BMPs. BMP9 (also known as growth differentiation factor 2, or GDF2) was originally identified from fetal mouse liver [25], and its possible roles include regulating the cholinergic phenotype of embryonic basal forebrain cholinergic neurons [26], inducing hematopoietic progenitor cell generation and colony formation [27], and maintaining homeostasis of iron metabolism [28]. In previous studies, it has been demonstrated that BMP9 is one of growth factors capable of significally inducing osteogenic differentiation of MPCs[12,29], while many other factors participant in the procedure, such as Retinotic Acid [5], Hey1 [30] and TGFβ1 [31]. It is conceivable that some molecules may act synergistically or antagonistically to perturb BMP9-mediated osteogenic differentiation of MPCs.
During the last twenty years of CXCL12 have been focused on, the CXCL12 was originally defined as a chemotaxis regulator for stem cell homing and recruitment [32], and bone marrow formation [33]. Interestingly, high CXCL12 expression is linked with a more primitive uncommitted stage of preosteogenic differentiation [34]. Despite it, previous study has demonstrated that CXCL12 also directly played a role in regulating osteogenic differentiation of mesenchymal stem cells induced by BMP2 [23]. And the relation between CXCL12 and BMP9 is limited in the research of hereditary hemorrhagic telangiectasia (HHT) and vascular remodeling [35]. In this report, we investigate the effect of CXCL12/CXCR4 signal axis on BMP9-induced osteogenic differentiation of MPCs.
Previous studies [34] and our data first demonstrated that high levels of CXCL12 and CXCR4 expressed in the murine mesenchymal progenitor cell line C3H10T1/2. In the present study, we confirmed that CXCL12 and CXCR4 expressions were significantly down-regulated at the immediate early stage of BMP9 stimulation coinciding with the commitment of C3H10T1/2 cells to osteoblast lineage. The phenomenon is accordant with previous studies that CXCL12 expression depressed when the human mesenchymal stem cells were cultured in osteogenic medium containing dexamethasone [34], and when the C2C12 cells were stimulated with BMP2, respectively [22]. These results strongly demonstrated that CXCL12/CXCR4 is an important and direct target of the BMP9 osteogenic signaling pathway. We hypothesis that CXCL12/CXCR4 signal axis may be required for initiation of BMP9 mediated osteogenic differentiation until the proliferation and differentiation of C3H10T1/2 cells. It is conceivable that: 1) the expression of CXCL12/CXCR4 may plays an important role in the early and middle osteogenesis maintained by BMP9 instead of the later part; 2) C3H10T1/2 cells are able to express enough CXCL12 and CXCR4 to support osteogenic differentiation mediated by BMP9, but their expression gradually decreased accompanied by the osteogenic differentiation, that is, in this experiment induced by BMP9.
In the present reports, we study the osteogenic differentiation signaling pathway by detecting several markers for preosteoblasts. ALP is a marker of early differentiation of osteoblasts, which is secreted by bone matrix [36]. In the early experiment we found out that in the procedure of osteogenic atomization of C3H10T1/2 cells which mediated by BMP9, ALP started expressing from 3rd day, reach peak at 7th day and decreased from 10th day or so [37], there for the expression of 7th day is chosen in this experiment. OCN is vitamin K dependent calcium binding proteins which synthesized and secreted by ossify and mainly deposits in bone matrix. Osteocalcin is regarded as one of the symbols osteoblast differentiation to mineralization period for it mainly appeared in mineralization period [38]. Calcium salt is inorganic substance in bone matrix and is called bone salts as well. The formation of calcium salt is the symbol of differentiation and maturity of ossify [37]. In consequence, the deposit of OCN and calcium salt are regarded as the symbol of middle-advanced stage of ossify differentiation and are tested at 14th and 21st day.
When the expression of CXCR4 was blocked by PTX before the stimulation of BMP9, the BMP9-induced ALP activity of C3H10T1/2 cells depressed, and the addition of exogenous CXCL12 was not able to rescue it. It is well established that CXCL12 activate directional cell movement via PTX-sensitive Gαi protein-coupled receptor, CXCR4 [39, 40]. The results of Adsi-CXCR4 and AMD3100 treatment are also consistent with this notion, but the depressions were less than PTX. As PTX was a non-specific inhibitor to CXCR4, maybe CXCL12 participated in BMP9-induced ALP activity by combining with other G protein. The timing of perturbing CXCL12/CXCR4 signal axis to elicit a block of BMP9-induced osteogenesis in C3H10T1/2 cells appears to be crucial. We find that if the processing factors were added afterward of the stimulation of BMP9, they were almost ineffective in perturbing BMP9-induced ALP activity, but the ALP activity stimulated by BMP9 could not be affected no matter the sequence of perturbing with CXCL12/CXCR4 signal axis or stimulation with BMP9 in C3H10T1/2 cells.
In the present studies, the expression of markers of early and middle stages of osteogenic differentiation, ALP and OCN, were reduced by the blocking of CXCL12 and CXCR4 before the stimulation of BMP9. In contrast, both the expression of ALP and OCN were also up-regulated by the addition of exogenous CXCL12 prior to the stimulation of BMP9. Moreover, blocking CXCL12/CXCR4 signal axis before the addition of BMP9 also resulted in the suppressing of marker for later stage of osteogenic differentiation, calcium deposition. However, the addition of exogenous CXCL12 have almost no effect on the calcium deposition induced by BMP9, suggesting that CXCL12/CXCR4 signal axis is involved in the early and middle stages of BMP9-dependent osteoinduction instead of the late stage. Similar results can also be researched in the HS5 MPCs, which is human bone marrow stromal cell line. Taken together, these results suggest that the expression of CXCL12 and CXCR4 may decrease after osteogenic induction with BMP9 because CXCL12/CXCR4 is no longer required once the osteogenic differentiation pathway has been set in motion.
In this context, the earliest expression of transcription factors associated with osteogenic differentiation induced by BMP9 in C3H10T1/2 cells, such as RUNX2, OSX, Plzf and Dlx5 were detected to be reduced by the blocking of CXCL12/CXCR4 signal axis. Furthermore, when processing factors against CXCL12 or CXCR4 were added to C3H10T1/2 cells before the addition of BMP9, the phosphorylation of Smad1/5/8 and Erk1/2 were reduced, suggesting that CXCL12 is involved in the early stages of BMP9-induced osteogenic differentiation, and the involvement is by the signal of Smad and MAPK.
In conclusion, our results demonstrated an important regulatory role for CXCL12 and CXCR4 in BMP9-stimulated osteogenic differentiation of C3H10T1/2 MPCs for the first time. CXCL12/CXCR4 signaling appears to be a prerequisite for BMP9-dependent osteogenic signaling. This knowledge will expand our understanding about how BMP9 cross-talks with other signaling pathways to support bone repair and regeneration, and will contribute to the identification of effective bone regeneration therapeutics to treat skeletal diseases.
Acknowledgements
We would like to thank Dr. Tongchuan He in University of Chicago for the gift of pAdEasy-1 and pAdTrace-TO4 vectors. This work was supported by the National Natural Science Foundation of China (NSFC30872770), Natural Science Foundation Project of CQ CSTC (2011BB5131) and National Ministry of Education Foundation of China (KJ120327).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gregory CA, Prockop DJ, Spees JL. Non-hematopoietic bone marrow stem cells: molecular control of expansion and differentiation. Exp Cell Res. 2005;306:330-5
2. Sacchetti B, Funari A, Michienzi S. et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324-36
3. Chamberlain G, Fox J, Ashton B. et al. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739-49
4. Deng ZL, Sharff KA, Tang N. et al. Regulation of osteogenic differentiation during skeletal development. Front Biosci. 2008;13:2001-21
5. Zhang W, Deng ZL, Chen L. et al. Retinoic acids potentiate BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. PLoS One. 2010;5:e11917
6. Rastegar F, Shenaq D, Huang J. et al. Mesenchymal stem cells: Molecular characteristics and clinical applications. World J Stem Cells. 2010;2:67-80
7. Jiang Y, Jahagirdar BN, Reinhardt RL. et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2007;447:879-80
8. Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191-220
9. Wagner ER, Luther G, Zhu G. et al. Defective osteogenic differentiation in the development of osteosarcoma. Sarcoma. 2011;2011:325238
10. Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233-41
11. Boraiah S, Paul O, Hawkes D. et al. Complications of recombinant human BMP-2 for treating complex tibial plateau fractures: a preliminary report. Clin Orthop Relat Res. 2009;467:3257-62
12. Luu HH, Song WX, Luo X. et al. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res. 2007;25:665-77
13. Hogan BL.Bone morphogenetic proteins. multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580-94
14. Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50-83
15. Luther G, Wagner ER, Zhu G. et al. BMP-9 induced osteogenic differentiation of mesenchymal stem cells: molecular mechanism and therapeutic potential. Curr Gene Ther. 2011;11:229-40
16. Luo J, Tang M, Huang J. et al. TGFbeta/BMP type I receptors ALK1 and ALK2 are essential for BMP9-induced osteogenic signaling in mesenchymal stem cells. J Biol Chem. 2010;285:29588-98
17. Nagasawa T, Nakajima T, Tachibana K. et al. Molecular cloning and characterization of a murine pre-B-cell growth-stimulating factor/stromal cell-derived factor 1 receptor, a murine homolog of the human immunodeficiency virus 1 entry coreceptor fusin. Proc Natl Acad Sci USA. 1996;93:14726-9
18. Blanchet X, Langer M, Weber C. et al. Touch of Chemokines. Front Immunol. 2012;3:175
19. Ratajczak MZ, Zuba-Surma E, Kucia M. et al. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915-24
20. Kucia M, Ratajczak J, Reca R. et al. Tissue-specific muscle, neural and liver stem/progenitor cells reside in the bone marrow, respond to an SDF-1 gradient and are mobilized into peripheral blood during stress and tissue injury. Blood Cells Mol Dis. 2004;32:52-7
21. Sun X, Cheng G, Hao M. et al. CXCL12 / CXCR4 / CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29:709-22
22. Zhu W, Boachie-Adjei O, Rawlins BA. et al. A novel regulatory role for stromal-derived factor-1 signaling in bone morphogenic protein-2 osteogenic differentiation of mesenchymal C2C12 cells. J Biol Chem. 2007;282:18676-85
23. Hosogane N, Huang Z, Rawlins BA. et al. Stromal derived factor-1 regulates bone morphogenetic protein 2-induced osteogenic differentiation of primary mesenchymal stem cells. Int J Biochem Cell Biol. 2010;42:1132-41
24. Xu DJ, Zhao YZ, Wang J. et al. Smads, p38 and ERK1/2 are involved in BMP9-induced osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. BMB Rep. 2012;45(4):247-52
25. Song JJ, Celeste AJ, Kong FM. et al. Bone morphogenetic protein-9 binds to liver cells and stimulates proliferation. Endocrinology. 1995;136:4293-7
26. López-Coviella I, Berse B, Krauss R. et al. Induction and maintenance of the neuronal cholinergic phenotype in the central nervous system by BMP-9. Science. 2000;289:313-6
27. Ploemacher RE, Engels LJ, Mayer AE. et al. Bone morphogenetic protein 9 is a potent synergistic factor for murine hemopoietic progenitor cell generation and colony formation in serum-free cultures. Leukemia. 1999;13:428-37
28. Truksa J, Peng H, Lee P. et al. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA. 2006;103:10289-93
29. Kang Q, Sun MH, Cheng H. et al. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11:1312-20
30. Sharff KA, Song WX, Luo X. et al. Hey1 basic helix-loop-helix protein plays an important role in mediating BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. J Biol Chem. 2009;284:649-59
31. Li RD, Deng ZL, Hu N. et al. Biphasic effects of TGFβ1 on BMP9-induced osteogenic differentiation of mesenchymal stem cells. BMB Rep. 2012;45:509-14
32. Wong D, Korz W. Translating an Antagonist of Chemokine Receptor CXCR4: from bench to bedside. Clin Cancer Res. 2008;14:7975-80
33. Sugiyama T, Kohara H, Noda M. et al. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977-88
34. Kortesidis A, Zannettino A, Isenmann S. et al. Stromal-derived factor-1 promotes the growth, survival, and development of human bone marrow stromal stem cells. Blood. 2005;105:3793-801
35. Young K, Conley B, Romero D. et al. BMP9 regulates endoglin-dependent chemokine responses in endothelial cells. Blood. 2012;120:4263-73
36. Fukuoka H, Shibata S, Suda N. et al. Bone morphogenetic protein rescues the lack of secondary cartilage in Runx2-deficient mice. J Anat. 2007;211:8-15
37. Zhao Dan, Luo Jinyong. The promotive effect of BMP9 in osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. Journal of Clinical and Experimental Medicine. 2011;10:1225-1230
38. Luegmayr E, Varga F, Glantschnig H. et al. 1, 25-Dihydroxy vitamin D3 and tri-iodothyronine stimulate the expression of a protein immunologically related to osteocalcin. J Histochem Cytochem. 1998;46:477-86
39. Poznansky MC, Olszak IT, Evans RH. et al. Thymocyte emigration is mediated by active movement away from stroma-derived factors. J Clin Invest. 2002;109:1101-1110
40. Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565-8
Author contact
![]() Corresponding author: Qiong Shi, Department of Laboratory Medicine, Chongqing Medical University, 1#, Yixueyuan Road, Chongqing, 400016, China. Tel: +86 023-68485239 Fax: +86 023-68485005 Email: anniesq8718com.
Corresponding author: Qiong Shi, Department of Laboratory Medicine, Chongqing Medical University, 1#, Yixueyuan Road, Chongqing, 400016, China. Tel: +86 023-68485239 Fax: +86 023-68485005 Email: anniesq8718com.

 Global reach, higher impact
Global reach, higher impact