Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(8):957-964. doi:10.7150/ijms.5632 This issue Cite
Research Paper
Inhibition of HCV 5′-NTR and Core Expression by a Small Hairpin RNA Delivered by a Histone Gene Carrier, HPhA
1. Phase I Clinical Trial Unit, First Hospital, Jilin University, Changchun 130021, China;
2. First Hospital, Jilin University, Changchun, China;
3. Cancer Hospital of Jilin Province, Changchun, China;
4. Key Laboratory for Molecular and Engineering of Ministry of Education, Jilin University, Changchun, China.
Received 2012-11-30; Accepted 2013-3-17; Published 2013-6-9
Abstract
siRNA (small interfering RNA) interference represents an exciting new technology that could have therapeutic applications for the treatment of viral infections. However, a major challenge in the use of siRNA as a therapeutic agent is the development of a suitable delivery system. We demonstrated that a new non-viral transgene carrier, recombinant archaeal histone from the hyperthermophile Pyrococcus horikoshii OT3 (HPhA), can transfect short hairpin RNA (shRNA) expressing plasmids into HL-7702 cells to inhibit the expression of HCV 5'NTR and Core protein and mRNA. Plasmids Psilencirle transfected by HPhA inhibited the expression of HCV 5'-NTR and Core protein and mRNA in HL-7702 cells. The transfection efficiency of HPhA in HL-7702 cells was not affected by 10% fetal calf serum (FCS). HPhA exhibited effects of transfection without apparent toxicity, and with high affinity for DNA. This suggests that HPhA may be useful for RNAi-based gene therapy in vivo.
Keywords: RNA interference, hepatitis C virus, small hairpin RNA, gene therapy, HPhA
Introduction
Hepatitis C virus (HCV) is a major cause of chronic liver disease and affects over 170 million individuals worldwide. Persistent HCV infection is a leading cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma [1-4]. The current standard of care for genotype 1 HCV, triple therapy of PEG-interferon, ribavirin, and a protease inhibitor, results in sustained virological response rates of 70-75%[2,4]. HCV belongs to the family Flaviviridae, and its genome is a positive-strand 9.6 kb RNA. HCV has a 5′-non-translated region (NTR), a long open reading frame, and a 3′-NTR. An internal ribosome entry site (IRES), containing the 5′-NTR and part of the Core coding region, forms a secondary structure and supports translation initiation of an HCV genome in a cap-independent manner [5]. HCV 5'-NTR and Core are conserved regions in the HCV genome [6-8].
RNAi is the process whereby double-stranded RNA (dsRNA) induces the sequence-specific degradation of homologous messenger RNA (mRNA) [9]. Introduction of siRNA into mammalian cells leads to mRNA degradation with exquisite sequence specificity without activating an interferon response. Thus, RNA interference represents an exciting new technology that could have therapeutic applications for the treatment of viral infections [10,11]. To date, RNAi has been used effectively to inhibit the replication of several different pathogenic human viruses in culture, including respiratory syncytial virus (RSV), poliovirus, papillomavirus, HIV-1, hepatitis C virus (HCV), and HBV (hepatitis B virus). Unfortunately, small RNAs have not been applied in treating diseases. One of the reasons is that there is no suitable delivery system for effectively transferring siRNA in vivo [10-15]. A major challenge in the use of siRNA (small interfering RNA) as a therapeutic agent is the development of a suitable delivery system. Because the main goal of in vivo delivery is to have active siRNA oligos in the target cells, the stability of siRNA oligos in the extracellular and intracellular environments after systemic administration is the most challenging issue [16].
The first hurdle is the size of the 21-nucleotide double-stranded siRNA oligos: these oligos are relatively small, and thus, are rapidly excreted through urine when administrated into the blood stream, even if siRNA molecules remain stable through chemical modifications. Second, the double-stranded siRNA oligos are relatively unstable in the serum environment and they can be degraded by RNase activity within a short period of time.
More recently, DNA expression vectors have been developed to express small hairpin RNAs (shRNA). These vectors employ the type III class of RNA polymerase promoters to drive the expression of siRNA molecules within cells [5,17,18]. DNA is relatively stable, but still can be degraded by DNase and it is difficult to introduce naked plasmids into cells. Therefore, suitable carriers are required for this purpose. Currently, there are two major methods by which siRNAs are delivered into hepatocytes in experimental animals [19,20]. One is the hydrodynamic method, in which siRNA expressing plasmids in a solution that is injected into the blood by the tail vein. This solution causes high pressure in blood circulation so that siRNA expressing plasmids enter into hepatocytes. The stability and efficiency of this method have been variable. Another method is the use of viral vectors. Their transfection efficiency is high, but there are many disadvantages such as immune response, integration, and mutations which pose obstacles for clinical use. Many non-viral carriers have been used in the delivery of DNA, which has a great advantage for its application in drug target validation and also allows multiple administrations of siRNA, which are crucial for siRNA therapeutic applications [21-24].
Several proteins as non-viral vectors have been studied using positive charge of the protein to bind with DNA to form complexes in cells [25]. Histone is a potential protein because of its excellent DNA-assembling ability. Our idea was to use HPhA which comes from the hyperthermophile Pyrococcus horikoshii OT3 which has the same basic structure of eukaryotic histones. Its molecular weight is smaller than eukaryotic histones, and it has good stability and DNA-assembling ability. In this paper, the aim was to investigate the possibility of using HPhA as carriers for transferring shRNA plasmids into cells to inhibit the expression of HCV 5′-NTR and Core protein.
Materials and Methods
Cells, HCV Replicon and carrier
HL-7702 cells were maintained at 37°C in an atmosphere of 5% CO2 in 1640 medium (Invitrogen) containing 20% fetal bovine serum, 200 U/ml of penicillin G, and 200 g/ml of streptomycin. HCV subgenomic replicon (genotype 1a) pCMV/T7 NTRCΔ-luc consisting of HCV 5′-NTR, part of Core sequences and a luciferase reporter gene, was kept in our laboratory. HphA was kindly provided by the Key Laboratory for Molecular and Engineering of Ministry of Education at Jilin University. Lipofectamine TM2000 (1 mg/mL), cationic liposomes were obtained from Invitrogen and used according to the manufacturer's recommendations.
shRNA expression constructs
The GenBank database was searched for unique sequences within HCV to exclude known cellular genes. Target sequences for the siRNAs were determined by using the Ambion web-based criteria, followed by generation of the HCV- specific siRNAs using the SilenCircleTM RNAi Kit (Allele Biotechnology & Pharmaceutical. Inc). Three sites, HCV -72 (5-aaagcgtctagccatggcgtt-3) and HCV -274 (5-aaaggccttgtggtactgcct-3) in the 5′NTR and HCV-365 (5-aagaaaaaccaaacgtaacaccaacc-3) in the Core of the common sequences of the HCV 1a. Five DNA inserts were designed to generate shRNA: HCV-72 sense 5'-acaccagcgtctagccatggcgttttgcttgaaaacgccatggctagacgctt-3'; antisense 3'-g tcgcagatcggtaccgcaaaacgaacttttgcggtaccgatctgcg a aaaaa-5'. HCV-274: sense 5'-acaccaggccttgtggtactgcctttgcttgaaaggcagtaccacaaggcctt-3'; antisense: 3'-g tccggaacaccatgacggaaacgaactttccgtcatggtgttccgga aaaaa-5'. HCV-365: sense: 5'-acaccaccaaacgtaacaccaaccttgcttgaaggttggtgttacgtttggtt-3' antisense: 3'-g ccacgagcacatagtgtgcaacgaacttgcacactatgtgctcgtggaaaaa-5'. luciferase-1019: sense: 5'-acaccattgcttctggtggcgctcttgcttgaagagcgccaccagaagcaat t-3'; antisense: 3'-g taacgaagaccaccgcgagaacgaacttctcgcggtggtcttcgtta aaaaa-5'. scrambled control: sense; 5'-acaccggtgctcgtgtatcacacgttgcttgaacgtgtgatacacgagcacct-3'; antisense: 3'-g ccacgagcacatagtgtgcaacgaacttgcacactatgtgctcgtggaaaaa-5' [26,27].
shRNA oligonucleotides were designed to contain a sense strand of 19-nucleotide sequences (from the HCV genome or scrambled sequence, not matched with the HCV genome or the host genome), followed by a short spacer (TTG CTT GAA), the reverse complement of the sense strand, and five thymidines as an RNA polymerase III transcriptional stop signal. The resultant plasmids (psh-72, psh-274, psh-365, and psh-1019) were used in the experiments. The plasmids psh-72, psh-274, psh-365, and psh-1019 were named to correspond with their respective targets, HCV-72, HCV-274, HCV-365 and luciferase-1019. The constructs contained the 3'-end of the sense strand and the 5'-end of the antisense strand, connected by a 9-nucleotide loop sequence. Scrambled siRNA (control) cloned into the same vector was used as a negative control in all of the experiments.
Cytotoxicity assays
HL-7702 cells were inoculated onto a 96-well plate at a density of 1×105/mL in 200 μL per well and cultured for 24 h prior to HphA or Lipofectamine TM2000 treatment. HphA or Lipofectamine TM2000 was added at various concentrations and allowed to incubate for 24 h. HphA or Lipofectamine TM2000 dissolved in 40 μL PBS, was added to 160 μL 1640 medium containing 10% fetal bovine serum. The cytotoxicity was measured by the reduction of methylthiazol tetrazolium (MTT; Sigma, USA) observed in mitochondria at 24 h after the initial treatment. The inhibition rate was calculated for each experimental group by comparison with MTT results from non-treated (1640 medium alone) cells [27].
Transfection
5 × 104 HL-7702 cells (500 μL) were plated in 24-well cluster plates for measuring luciferase activity after transfection. The following day, the plasmids pCMV/T7 NTRCΔ-luc, 1 μg, and shRNA expression plasmids (psh-72, psh-274, psh-365, psh-1019 and scrambled control), 1 μg, were simultaneously introduced into HL-7702 cells by 3 μg HphA and 3 μL Lipofectamine TM2000 (Invitrogen), respectively [25].
1.5×106 HL-7702 cells (15 μL) were plated in 75 cm2 cell culture bottles for Western Blotting after transfection. The following day, the plasmids pCMV/T7 NTRCΔ-luc, 30 μg, and shRNA expression plasmids (psh-72, psh-274, psh-365, psh-1019 and scrambled control), 30 μg, were simultaneously introduced into HL-7702 cells by 90 μg HphA and 90 μL Lipofectamine TM2000 (Invitrogen), respectively.
HphA and Lipofectamine TM2000 were dissolved in medium containing 10% FCS or without 10% FCS when transfection. After transfection 6 h, the cell medium was exchanged for fresh medium containing 20% fetal bovine serum [27].
Western Blotting
Cells were harvested using sodium dodecyl sulfate sample buffer in 72 h after transfection. Proteins were subjected to electrophoresis on a 10% polyacrylamide gel and transferred onto a nitrocellulose membrane (Bio-Rad). The membrane was probed with a monoclonal antibody to 5′-NTR (Biodesign International, Saco, ME) or actin (Santa Cruz Biotechnology, Santa Cruz, CA). Proteins were visualized using an enhanced-chemiluminescence detection kit (Amersham Pharmacia, Piscataway, NJ) [11].
Luciferase activity measuring
Cells in 24-well cluster plates were digested by trypsogen 72 h after transfection. Cells were washed twice with PBS, and then 100 μl cell lysate was added. Cells were agitated for 15 sec, then centrifuged for 15 sec. Supernatant was transferred to a new tube and placed at room temperature, then 100 μl supernatant was added to 100 μl luciferase substrate per well in 96-well plates. Luciferase activity was measured by a FLX800 luminometer [11].
DNA binding by HphA
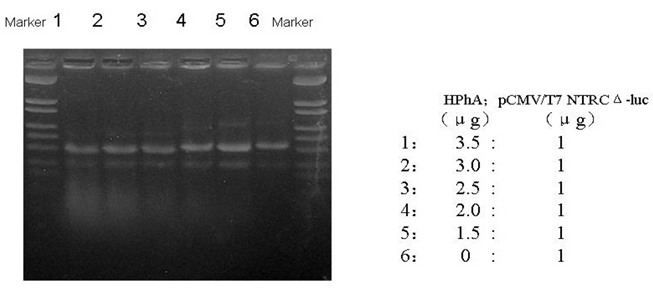
HphA was mixed with plasmids at ratios of 0 : 1, 1.5 : 1, 2.0 : 1, 2.5 : 1, 3.0 : 1, 3.5 : 1, and the mixture of HphA and DNA was electrophoresed for 12 h on an agarose gel (0.7 V/cm) [25].
Intracellular RNA purification and analysis
HL-7702 cells were harvested by trypsin digestion 72 h after transfection and washed three times with phosphate buffered saline (PBS, pH 7.3). Total RNA was isolated from cells using the TRIZOL reagent (Invitrogen). For RT-PCR analysis, RNA was reverse-transcribed at 42°C for 90 min using a commercially available cDNA synthesis kit (Invitrogen). PCR was performed with gene specific primers for HBV mRNA and β-actin: HCV (forward), 5′-TGCACCATGAGCACGAATCCTAAAC-3′; HCV nt2659-2636 (reverse), 5′-CCGGGAACTTAACGTCCTGTGGGCG-3′; β-actin (forward), 5′-ACGGCCAGGTCATCACCAT-3′; β-actin (reverse), 5′-AGGCTGGAAGAGTGCCTCAG-3′. qPCR was performed on the ABI 7300 Sequence Detection System using the SYBR Green kit (Invitrogen). The fold increase was calculated using 2-ΔΔCT [38].
Statistical analysis
Statistical analyses were performed on SAS software version 8.0 (USA). Data are expressed as mean ± standard deviation (SD). Simple comparisons between groups were carried out with the Student's t-test. A P-value less than 0.05 was considered statistically significant.
Results
Cytotoxicity assays
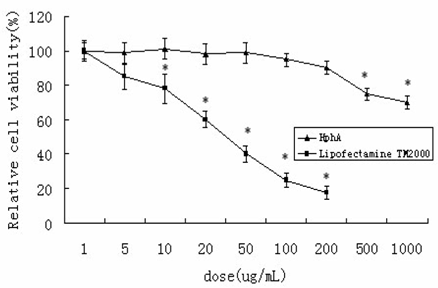
In order to evaluate toxicity, the number of surviving HL-7702 cells was monitored after 24 h incubation with various concentrations of either the HPhA or Lipofectamine TM2000. No apparent cytotoxicity was observed after treatment with HPhA at lower concentrations (200 μg/mL), whereas incubation with even lower amounts of Lipofectamine TM2000 caused considerable cell death (10 μg/mL). This finding suggested that the suppressive activities of HPhA on expression of HCV 5′-NTR and Core were not a result of induced cytotoxicity (Figure 1).
HphA binding DNA
To determine the ability of HphA to bind plasmids, HphA was mixed with plasmids in various ratios, and agarose gel electrophoresis was performed. The results showed that when HphA bound DNA the mixture moved faster in an agarose gel (Figure 2).
Cytotoxicity of LipofectamineTM2000 or HPhA in HL-7702 cells. The data represent mean±SD (n=4). *P<0.05 vs. the corresponding non-treated control (Student's t-test).
An agarose gel showing HPhA binding of DNA.
Influence of serum on transfection
HCV subgenomic replicon pCMV/T7 NTRCΔ-luc was transfected into HL-7702 cells by LipofectamineTM2000 or HPhA in medium with or without FCS. Luciferase activity was measured 72 h after transfection. They were 57±49, 4468±223, 4950±359 and 4538±268 RFU respectively at LipofectamineTM2000 With FCS (10%), LipofectamineTM2000 Without FCS, HphA With FCS (10%) and HphA Without FCS group in medium. Luciferase activity was lower at LipofectamineTM2000 With FCS (10%) than it in the other group in medium (p<0.05). In medium containing 10% FCS, no expression of luciferase was observed when LipofectamineTM2000 was used to transfect plasmids pCMV/T7 NTRCΔ-luc into cells. However, expression of luciferase was clearly shown when HphA was used to transfect.
Specific shRNA introduced by HphA inhibited the expression of HCV 5′NTR and Core
The plasmid pCMV/T7 NTRCΔ-luc and shRNA expression plasmids (psh-72, psh-274, psh-365, psh-1019 and scrambled control) were simultaneously introduced into HL-7702 cells (co-transfection) by HphA. The results showed that specific shRNA introduced by HphA could obviously inhibit the expression of HCV 5′NTR and Core in medium containing 10% FCS (59%, 59%, 55%) and in medium without FCS (58%, 62%, 54%), p<0.05. The luciferase activity was the same with and without FCS in the medium for the same shRNA group, p>0.05. The inhibition rates were different for the various targeting sites (Table 1).
Specific shRNA introduced by HphA inhibits the expression of HCV 5′-NTR and Core by Western Blot
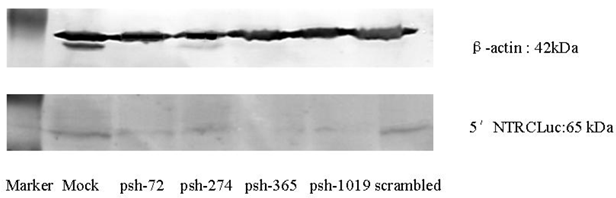
Western Blot showed that the expression after inhibition of the HCV 5′-NTR was lower for the shRNA expression plasmids (psh-72, psh-274, psh-365, psh-1019) group than the Mock and scrambled groups (Figure 3).
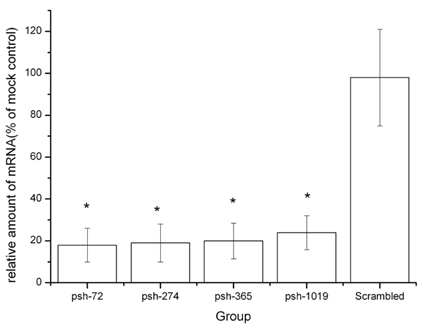
To characterize the functional effects on mRNA by HCV-specific knock-down, we measured the amounts of HCV mRNA present after exposure to shRNA. Intracellular HCV mRNA levels were analyzed by real time PCR. As shown in Figure 4, the shRNA expression plasmids (psh-72, psh-274, psh-365, psh-1019) reduced the levels of HCV mRNA in all groups. The inhibition rates for plasmids psh-72, psh-274, psh-365, psh-1019 were significantly higher than that for mock and scrambled groups (P<0.05). There were no significant differences between levels in mock and scrambled groups (p=0.81). The psh-72 plasmid yielded the highest inhibition rate (82%) of mRNA levels.
We also studied an HCV replicon Huh-7 cell line kindly provided by the Department of Hepatology of the First Hospital, JiLin University. This replicon contains a full-length genome of an HCV genotype 1a virus. The shRNA expression plasmids (psh-72, psh-274, psh-365, psh-1019) reduced the levels of HCV RNA in all groups. The inhibition rates (%) for plasmids psh-72, psh-274, psh-365, psh-1019 were 85±8.5, 88.5±7.4, 90±9.8, 87±9.5 respectively, which were significantly higher than that for mock (control) and scrambled (3.7±3.6) groups (P<0.05). There were no significant differences between levels in mock and scrambled groups (P=0.83).
Specific shRNA Introduced by HphA Inhibits the Expression of HCV 5′-NTR and Core Genes
| Luciferase activity (RFU) | Blank | Mock | Psh-72 | Psh-274 | Psh-365 | Psh-1019 | Scrambled |
|---|---|---|---|---|---|---|---|
| With FCS (10%) in medium | 0 | 6154 5224 6157 | 2095 3305 2608 | 2458 1987 2509 | 3257 1926 2425 | 2982 2334 731 | 5471 6234 5012 |
| 5845±414 | 2269±424* | 2318±221* | 2536±481* | 2016±856* | 5572±441 | ||
| 59% | 59% | 55% | 64% | ||||
| Without FCS in medium | 0 | 5843 5709 5324 | 2178 2357 2987 | 2475 2067 2378 | 2878 2390 2641 | 2309 2256 1668 | 5617 6146 5987 |
| 5625±200 | 2507±319* | 2307±160* | 2635±164* | 2077±273* | 5917±199 | ||
| Inhibition rate | 58% | 62% | 54% | 65% |
* p<0.05 compared with luciferase activity in the Mock group in the same FCS medium
The effects of shRNA introduced by HphA on the expression of HCV 5′-NTR and Core.
The effects of shRNA introduced by HphA on the expression of HCV mRNA. *: p<0.05 VS scrambled group.
Discussion
Wilson et al. reported that specific siRNA targeting to NS3 and NS5 of the HCV genome could inhibit replication and expression of HCV replicons. However, after repeated RNAi therapy, they found that resistance to the siRNAs developed, and the siRNAs targeting to the other sites were still active [28]. The sequence analysis of NS3 and NS5 showed that there were mutations in targeting sequences of siRNA. After multiple exposures to RNAi therapy, increasing numbers of mutations have been found. This development of resistance to siRNAs is important, and may suggest that viruses mutate under the pressure of siRNA.
Two methods could prevent resistance due to virus mutation: 1. targeting sites should be chosen in the conservative areas; for example, 5'-NTR and Core region of HCV in our study; 2. the use of several different siRNAs targeting sites simultaneously, could limit the development of escape mutation of viruses. This is also an important advantage of siRNAs. The siRNAs prepared by endonuclease (esiRNAs) has been used to simultaneously target several sites of the viral genome with excellent effects [27].
Histones are the basic component of eukaryotic chromatin. Almost all of DNA binds to histones and folds to form chromatin fibers. In eukaryotes, histones and DNA form nuclear bodies which are the basic structural units of chromatin. The core of the nuclear body is histone, wound tightly by DNA. However, the tight binding can loosen during DNA binding to other proteins such as polymerase [25,29,30].
The amino acid sequence of HphA (60-70 amino acids) is similar to eukaryotic histones, but it is smaller (approximately 100~140 amino acids). HphA is the core structure of eukaryotic histone [31]. The complete genomic sequence of Pyrococcus horikoshii OT3 has been reported. It predicted open reading frames PHS051 and PHS046 [32,33]. KEY Laboratory for Molecular and Engineering of Ministry of Education in Jilin University prepared this recombinant archaeal histone HphA and used it for transfection in NIH 3T3, HEK 293, HL-7702, HepG2 and Cos 7 cell lines in vitro. HphA maintains basic second and third class structure of a eukaryotic histone, its molecular weight is less (70 amino acids) and it is stable, thermotolerant, and has good DNA-assembling ability [32].
We mixed HphA and siRNA expressing plasmids in various ratios, and then the plasmid DNA-HPhA complexes were electrophoresed for 12 hours in an agarose gel [34]. The current result showed that the mixture moved faster in gel. It suggested that HphA bound to plasmids changed the size or charge of the complexes resulting in faster migration.
Cell toxicity was evident with shrinkage and cell debris after transfection by LipofectamineTM2000 and cell death. Cells transfected by HphA were healthy after 20 h, even without replacement of the culture medium. The data suggested that HphA is non-toxic and safe.
The influence of serum on transfection should be considered in vivo, because even if transfection is very successful in vitro, it may not be successful in vivo due to serum effects. Sometimes the effect of transfection is much lower in vivo [35].
Therefore, we investigated the effect of transfection of HphA and LipofectamineTM2000 in the presence of serum [36]. Serum components such as serum albumin, immunoglobulin, fibrin and apoprotein, could bind with liposomes and alter the charge on the surface of liposomes. This could decrease the effectiveness of liposome-mediated transfection [37].
In summary, HphA has good stability and DNA-assembling ability as a non-viral transgene carrier. The transfection mediated by HphA was not affected by FCS. HPhA may be useful for RNAi-based gene therapy in vivo, but this will require further study for confirmation.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number: 30872174).
Competing Interests
The authors have declared that no competing interest exists.
References
1. http://www.who.int/mediacentre/factsheets/fs164/en/index.html
2. Jeffers L. Hepatocellular carcinoma: an emerging problem with hepatitis C. J Natl Med Assoc. 2000;92:369-371
3. Michielsen P, Ho E, Francque S. Does antiviral therapy reduce the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Minerva Gastroenterol Dietol. 2012;58:65-79
4. Qu LS, Chen H, Kuai XL. et al. Effects of interferon therapy on development of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis: A meta-analysis of randomized controlled trials. Hepatol Res. 2012;42:782-9
5. Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497-500
6. Choo QL, Kuo G, Weiner AJ. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362
7. Chuang WL, Yu ML, Dai CY. et al. Treatment of chronic hepatitis C in southern Taiwan. Intervirology. 2006;49:99-106
8. Yu ML, Chuang WL. Treatment of chronic hepatitis C in Asia: when East meets West. J Gastroenterol Hepatol. 2009;24:336-345
9. Lindenbach BD, Rice CM. RNAi targeting an animal virus: news from the front. Mol Cell. 2002;9:925-927
10. Randall G, Rice CM. Interfering with hepatitis C virus RNA replication. Virus Res. 2004;102:19-25
11. Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050-7058
12. Alvarez R, Elbashir S, Borland T. et al. RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob Agents Chemother. 2009;53:3952-3962
13. Boden D, Pusch O, Lee F. et al. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531-11535
14. McCaffrey AP. RNA interference inhibitors of hepatitis B virus. Ann N Y Acad Sci. 2009;1175:15-23
15. Yoshinouchi M, Yamada T, Kizaki M. et al. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by E6 siRNA. Mol Ther. 2003;8:762-768
16. Lapierre J, Salomon W, Cardia J. et al. Potent and systematic RNAi mediated silencing with single oligonucleotide compounds. RNA. 2011;17:1032-1037
17. Sui G, Soohoo C, Affar el B. et al. (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:5515-5520
18. Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:6047-6052
19. Rychahou PG, Evers BM. Hydrodynamic delivery protocols. Methods Mol Biol. 2010;623:189-195
20. Wesche-Soldato DE, Lomas-Neira J, Perl M. et al. Hydrodynamic delivery of siRNA in a mouse model of sepsis. Methods Mol Biol. 2008;442:67-73
21. Gao K, Huang L. Nonviral methods for siRNA delivery. Mol Pharm. 2009;6:651-658
22. Lu PY, Xie F, Woodle MC. In vivo application of RNA interference: from functional genomics to therapeutics. Adv Genet. 2005;54:117-142
23. Sun Y, Li Z, Li L, Li J, Liu X. et al. Effective inhibition of hepatitis B virus replication by small interfering RNAs expressed from human foamy virus vectors. Int J Mol Med. 2007;19:705-711
24. Yang FF, Huang W, Li YF. et al. Current status of non-viral vectors for siRNA delivery. Yao Xue Xue Bao. 2011;46:1436-1443
25. Li YY. Immunogenicity of the recombinant archaeal histone as a non-viral gene delivery. immunological journal. 2007;5:474-479
26. Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57-67
27. Kronke J, Kittler R, Buchholz F. et al. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J Virol. 2004;78:3436-3446
28. Wilson JA, Jayasena S, Khvorova A. et al. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci U S A. 2003;100:2783-2788
29. Kaouass M, Beaulieu R, Balicki D. Histonefection: Novel and potent non-viral gene delivery. J Control Release. 2006;113:245-254
30. Li YY, Wang R, Zhang GL. et al. An archaeal histone-like protein mediates efficient p53 gene transfer and facilitates its anti-cancer effect in vitro and in vivo. Cancer Gene Ther. 2007;14:968-975
31. Mahato RI. Non-viral peptide-based approaches to gene delivery. J Drug Target. 1999;7:249-268
32. Weng L. Cloning, Expression, Characterization and Application in Gene Therapy of Histone from Hyperthermophilic Archaeon Pyrococcus Horikoshii OT3. Jilin, China: Jilin university. 2004
33. Yao J, Kathpalia P, Bushinsky DA. et al. Hyperresponsiveness of vitamin D receptor gene expression to 1,25-dihydroxyvitamin D3. A new characteristic of genetic hypercalciuric stone-forming rats. J Clin Invest. 1998;101:2223-2232
34. Wu GY, Wu CH. Receptor-mediated gene delivery and expression in vivo. J Biol Chem. 1988;263:14621-14624
35. Kim YJ, Kim TW, Chung H. et al. The effects of serum on the stability and the transfection activity of the cationic lipid emulsion with various oils. Int J Pharm. 2003;252:241-252
36. Golan S, Aytar BS, Muller JP. et al. Influence of biological media on the structure and behavior of ferrocene-containing cationic lipid/DNA complexes used for DNA delivery. Langmuir. 2011;27:6615-6621
37. Cherng JY, van de Wetering P, Talsma H. et al. Effect of size and serum proteins on transfection efficiency of poly ((2-dimethylamino)ethyl methacrylate)-plasmid nanoparticles. Pharm Res. 1996;13:1038-1042
38. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8
Author contact
![]() Corresponding author: Dr. Junqi Niu, Department of Hepatology, First Hospital, Jilin University, Changchun 130021, China; Tel/Fax: +86-431-85612708; Email: hzhang09jlu.edu.cn
Corresponding author: Dr. Junqi Niu, Department of Hepatology, First Hospital, Jilin University, Changchun 130021, China; Tel/Fax: +86-431-85612708; Email: hzhang09jlu.edu.cn

 Global reach, higher impact
Global reach, higher impact