Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(8):948-956. doi:10.7150/ijms.5642 This issue Cite
Research Paper
Down-Regulated CK8 Expression in Human Intervertebral Disc Degeneration
1. Department of Orthopaedics, Xijing Hospital, Fourth Military Medical University, 127 Changle Western Road, Xi'an, P. R. China.
2. Aerospace Medical School, 15 Changle Western Road, Fourth Military Medical University, Xi'an, P. R. China.
3. Department of Orthopaedics and Traumatology, University of Hong Kong, Pokfulam, Hong Kong, SAR China.
† These authors contribute equally to this work.
Received 2012-12-4; Accepted 2013-5-24; Published 2013-6-8
Abstract
As an intermediate filament protein, cytokeratin 8 (CK8) exerts multiple cellular functions. Moreover, it has been identified as a marker of notochord cells, which play essential roles in human nucleus pulposus (NP). However, the distribution of CK8 positive cells in human NP and their relationship with intervertebral disc degeneration (IDD) have not been clarified until now. Here, we found the percentage of CK8 positive cells in IDD (25.7±4.14%) was significantly lower than that in normal and scoliosis NP (51.9±9.73% and 47.8±5.51%, respectively, p<0.05). Western blotting and qRT-PCR results confirmed the down-regulation of CK8 expression in IDD on both of protein and mRNA levels. Furthermore, approximately 37.4% of cell clusters were CK8 positive in IDD. Taken together, this is the first study to show a down-regulated CK8 expression and the percentage of CK8 positive cell clusters in IDD based upon multiple lines of evidence. Consequently, CK8 positive cells might be considered as a potential option in the development of cellular treatment strategies for NP repair.
Keywords: intervertebral disc degeneration, nucleus pulposus, cytokeratin 8, cell clusters, Immunofluorescence, qRT-PCR.
Introduction
Intervertebral disc degeneration (IDD) is implicated as a vital cause of low back pain (LBP), which results in not only great individual suffering, but global socioeconomic burden [1-3]. It has been estimated that approximately 80% of the population might have LBP at some point in their lives, leading IDD to an important public health issue [4]. The aetiology of IDD has been ascribed to various factors, including mechanical stress [5,6], cell death [7,8], gene polymorphisms [9,10], aging [11,12] and aberrant expression of microRNAs [7]. Despite intensive research activities in this field, the pathophysiology of IDD is still not well explored.
The intervertebral disc is comprised of a central gelatinous nucleus pulposus (NP) surrounded by annulus fibrosus (AF) and sandwiched by cartilage endplate. The normal human NP consists of extracellular matrix interspersed by a small number of cells that make up about 1% of the total volume [13]. Accumulating evidence has shown notochord cells play essential roles in the development and function of human NP [14-18]. Moreover, in vitro experiments have demonstrated that notochord cell is a vital factor to regulate extracellular matrix production and cell proliferation in some adult mammals [14,19-21]. However, notochord cells disappear in the first decade of human being and thereafter are replaced by chondrocyte-like cells with the occurrence of IDD [17,20,22].
Amongst the proteins of notochord cells, cytokeratin 8 (CK8) has been identified as a classical marker [23-26]. CK8 is a member of the cytokeratins family which belongs to the intermediate filament proteins of epithelial cells including notochord cells of NP [27]. Meanwhile, cytokeratins exert multiple functions due to their unique structural feature, i.e., the maintenance of response to mechanical stress, the regulation of Fas-mediated apoptosis and the modulation of cell size and protein synthesis. Stosiek et al. identified the co-expression of CK8 and vimentin in adult human NP, which indicated a close relationship between these cells and notochord cells [26]. Weiler et al. noted that a substantial variation of CK8 positive cells presence was linked with age in human disc [23]. As for bovine NP cell makers, Gilson et al. found that about 10% of NP cells were CK8 positive and appeared as small isolated clusters surrounded by gelatinous matrix [28].
However, the distribution of CK8 positive cells in human NP and their relationship with IDD have not been clarified until now. Here, we assumed that CK8 might be closely linked with notochord cells and the maintenance of normal human NP. Accordingly, this study was aimed to address the expression of CK8 in human NP and its relationship with IDD using immunofluorescence staining, western blotting and qRT-PCR. Meanwhile, the relationship between cell clusters and CK8 positive cells was also studied.
Materials and Methods
The study was approved by the institutional ethics review board (No. 20111103-7) with written informed consents obtained from each patient or his/her relatives.
Donor grouping
The study groups consisted of NP from cadavers and patients as below: Normal control group from 8 cadavers graded as Grade I (age 31.6±7.4years, range 21-42 years), disease control group from 9 patients with scoliosis undergoing anterior discectomy and fusion graded as Grade II [45] (age 20.1±2.1 years, range 16-24 years) and IDD group from 17 patients undergoing discectomy (age 32.2±7.1years, range 23-46years). Intervertebral discs were graded according to Pfirrmann's grading system [29].
NP specimen preparation
NP specimens were obtained from cadavers, patients with scoliosis and IDD as normal control, disease control and IDD samples, respectively. As for the cadavers, magnetic resonance imaging (MRI) data before death in their recent health examination records were collected. Patients with spinal stenosis, tumors, infections, and previous lumbar disc surgery within 3 months were excluded from the study.
Human NP cells isolation and culture
All specimens were obtained within 1 hour either after autopsy or surgery. Subsequently, the NP was separated from the AF using a stereotaxic microscope carefully and washed with Hank balance salt solution to eliminate contamination and blood. Specimens were digested for 40 minutes in 0.2% pronase, then washed with Hank balance salt solution and incubated in 0.25% type II collagenase at 37°C under gentle agitation. After 4 hours, remaining tissue debris was removed through a 40 µm cell strainer. Cells were centrifuged at 200 g for 8 min and seeded in culture flasks cultured with F-12 medium (containing 10% FBS, 1% P/S) in 5% CO2 and 20% oxygen incubator. Cells cultured for 3 days were used in the following western blotting and qRT-PCR steps.
Immunofluorescence staining of frozen specimens
Specimens were snap-frozen and cut in 25μm transversally with a cryostat microtome. The frozen sections were fixed in methanol at -20°C for 30 min. Before treatment with the primary antibody, nonspecific binding was blocked by treatment with 1% bovine serum albumin phosphate-buffered saline (PBS), then the samples were incubated in the rabbit monoclonal antibody to CK8 (5 μg/mL) (ab32357, Abcam, Cambridge, USA) in 12h in 4°C. The samples were washed in PBS for 5 min and incubated with Alexa 488-conjugated goat anti-rabbit secondary antibodies (Molecular Probes, Eugene, OR, USA) for 30 min in the dark at room temperature. For DNA counterstain, the samples were incubated in medium containing DAPI (4'-6-diamid-ino-2-phenylindole) after washed with PBS. Slides were visualized using a Leica microscope (Leica, Wetzlar, Germany).
For all the frozen specimen strips, 10 fields in 20X were selected randomly. Quantitative analysis was performed by two pathologists who were unaware of the clinical data. The positively stained cells and cell clusters were counted as a fraction of the area. Cell clusters were defined as aggregates of three or more cell nucleus in the DAPI counter-stain view. The percentage of CK8 positive cells of all specimens and stained cell clusters were calculated, respectively. Whole cohort of samples in Table 1 was used for the data analysis.
Summary of demographic data
| Patients NO. | Age | Gender | Level | Degree* | Total NP Cells /10PFs | Total CK8 positive cells/10PFs | Percentage of total CK8 positive cells (%) | CK8 positive clusters/all clusters (%) |
|---|---|---|---|---|---|---|---|---|
| Normal control | ||||||||
| 1 | 42 | M | L4/5 | I | 545 | 286 | 52.5 | |
| 2 | 25 | M | L4/5 | I | 578 | 327 | 56.6 | |
| 3 | 33 | F | L4/5 | I | 439 | 175 | 40 | |
| 4 | 27 | M | L4/5 | I | 365 | 243 | 66.7 | |
| 5 | 40 | M | L4/5 | I | 451 | 276 | 61.2 | |
| 6 | 39 | F | L4/5 | I | 323 | 133 | 41.2 | |
| 7 | 21 | M | L4/5 | I | 570 | 306 | 53.7 | |
| 8 | 26 | M | L4/5 | I | 641 | 277 | 43.2 | |
| Disease control | ||||||||
| 9 | 17 | F | L1/2 | II | 305 | 148 | 48.5 | 45.6 |
| 10 | 20 | F | L1/2 | II | 477 | 223 | 46.8 | 32.1 |
| 11 | 24 | F | L1/2 | II | 472 | 235 | 49.8 | 43.8 |
| 12 | 23 | F | L1/2 | II | 543 | 302 | 55.6 | 24.9 |
| 13 | 19 | F | L1/2 | II | 521 | 209 | 40.1 | 35.0 |
| 14 | 18 | F | L1/2 | II | 483 | 254 | 52.6 | 39.8 |
| 15 | 16 | M | L1/2 | II | 296 | 127 | 42.3 | 41.2 |
| 16 | 19 | M | L1/2 | II | 563 | 304 | 54 | 29.3 |
| 17 | 20 | M | L1/2 | II | 432 | 174 | 40.3 | 35.1 |
| IDD | ||||||||
| 18 | 32 | M | L4/5 | V | 458 | 136 | 29.7 | 42.7 |
| 19 | 23 | M | L4/5 | IV | 324 | 102 | 31.5 | 34 |
| 20 | 41 | M | L4/5 | V | 422 | 108 | 25.6 | 27.5 |
| 21 | 33 | F | L4/5 | IV | 532 | 156 | 29.3 | 56.4 |
| 22 | 34 | M | L4/5 | V | 418 | 78 | 18.7 | 30.3 |
| 23 | 28 | F | L5/S1 | IV | 356 | 93 | 26.1 | 17.8 |
| 24 | 29 | F | L5/S1 | IV | 462 | 139 | 30.1 | 63.3 |
| 25 | 24 | M | L4/5 | IV | 382 | 78 | 20.4 | 43.6 |
| 26 | 46 | M | L4/5 | IV | 532 | 147 | 27.6 | 31.8 |
| 27 | 26 | F | L5/S1 | V | 389 | 105 | 27 | 29 |
| 28 | 28 | M | L4/5 | IV | 463 | 93 | 20.1 | 19.4 |
| 29 | 42 | F | L5/S1 | IV | 386 | 87 | 22.5 | 53.1 |
PF = power field (X 20). * Pfirrmann's grading system. Normal control group was from cadavers, disease control group was from patients with scoliosis and IDD group was from patients with disc degeneration undergoing discectomy. CK8 positive clusters/all cell clusters (%): the number of clusters which were CK8 positive/the number of total clusters.
Double immunofluorescence of CK8 and CD24
Specimens were simultaneously incubated with rabbit monoclonal antibody to CK8 and mouse anti-human CD24 monoclonal antibody (catalog no. 555426; BD PharMingen, San Diego, CA) for 1 hour at room temperature. Following washed in PBS for 5 min, specimens were incubated with Alexa 488-conjugated goat anti-rabbit secondary antibody and 594 fluor-conjugated anti-mouse secondary antibody (Abcam, Cambridge, USA) for 30 min in the dark at room temperature. Slides were visualized using a Leica microscope (Leica, Wetzlar, Germany).
Western blotting
NP cells were cultured and trypsinized. Following washed with PBS, cell lysates were prepared in MOPS buffer on ice for 30 min. Debris of the cells was removed by centrifugation for 20 min at 4°C. The protein concentration was measured by the BCA assay (Sigma, Saint Louis, USA). Following electrophoresized in 10% Bis-Tris gel, equal amounts of 15ug proteins were transfered to PVDF membrane (0.45 mm). Then samples were blocked in the nonspecific binding sites overnight with 3% skim milk TBST (50 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween 20). Subsequently, the membranes were incubated for 1 h at room temperature with rabbit monoclonal antibody specific to CK8 (Abcam, Cambridge, USA, 1:1000), and mouse monoclonal antibody specific to β-actin (Sigma, Saint Louis, USA, 1:2000) as control. Antibody labeling was identified using goat anti-rabbit or anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology, Boston, USA 1:10000), Membranes were treated with ECL Plus according to the manufacturer's instructions (Amersham Pharmacia Biotech, Umea, Sweden). Images were analyzed by densitometry from Scion Image. To measure the amount of CK8 and β-actin expression, the CK8/β-actin ratio was calculated for each sample. Then, the relative expression of CK8 was obtained. NP cells were obtained from first eight individuals of each group in Table 1.
qRT- PCR
Total RNA in NP cells was isolated using TRIzol® Reagent (Ambion Invitrogen, Carlsbad, CA, USA) according to the manufacturer´s protocol. Reverse transcription to cDNA was performed using a High-Capacity cDNA Archive Kit (ABI, Foster City, CA, USA). Predesigned primers for human CK8 and human GAPDH as control were designed using OligoPerfectTM Designer Software (Invitrogen) and purchased from Sangon (Sangon, Shanghai, China). RNA concentrations were measured using a NanoDrop instrument (NanoDrop, Wilmington, DE, USA). The levels of CK8 mRNA were normalized to GAPDH mRNA controls. All RT reactions, including GAPDH controls, were run in triplicate in a GeneAmp PCR 9700 Thermocycler (ABI). Reverse transcriptions were performed at 37°C for 15 minutes and 85°C for 5 seconds. Quantitative real-time PCRs were done on a StepOne Plus device (Applied Biosystems) with SYBR Premix Ex Taq™ kit (TaKaRa). 1μg of cDNA was amplified at 95°C for 15 seconds followed by 40 cycles of 95°C, 5 seconds and 60°C for 30 seconds. The dissociation curves were run for all completed SYBR Green reactions to rule out non-specific amplifications and primer-dimers. The relative amounts of CK8 mRNA were calculated using the comparative Ct (2-ΔΔCt) method [30] and sample from the cadaver group was used as control sample. NP cells were obtained from first eight individuals of each group in Table 1. The primers used in the study were shown in Table 2.
Statistical analysis
Student's t-test was performed to compare two-group parameters (IDD and normal control, IDD and disease control). A p value less than 0.05 was considered as statistically significant. The SPSS statistical package (SPSS, Chicago, IL, USA) was used for the statistical analysis.
Results
The demography of individual's data was listed. Meanwhile, the percentage of total CK8 positive cells of each individual and the percentage of cell clusters with CK8 positive in each patient were shown (Table 1). In addition, data analysis showed that there was no significant difference about correlating CK8 expression versus donor age, which might own to the relatively small sample size in this study.
Expression of CK8 positive cells in NP
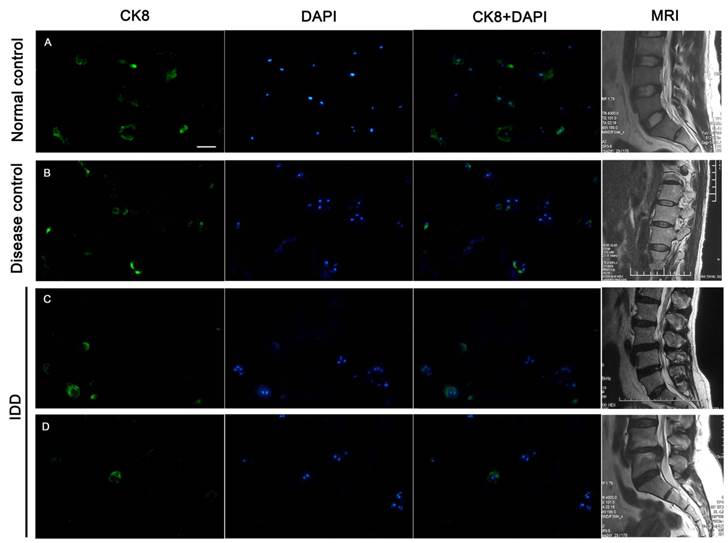
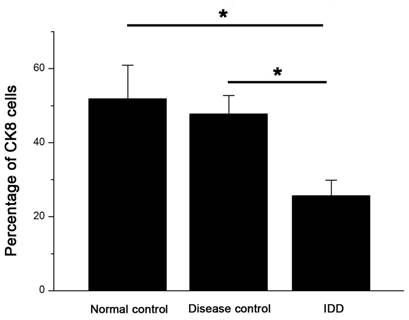
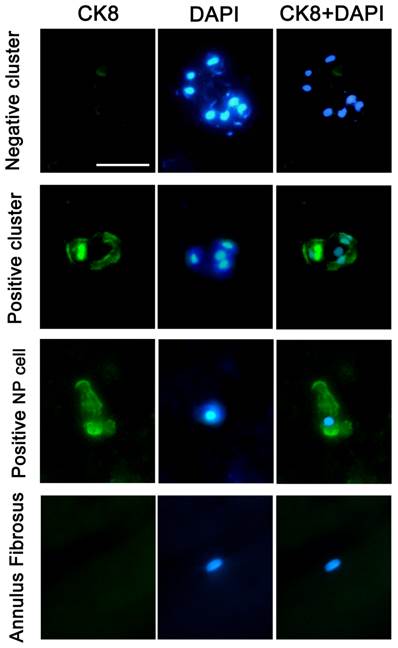
Immunofluorescence staining of frozen specimens showed a subset of CK8 positive cells in human NP (Figure 1).The percentage of CK8 positive cells in IDD (25.7±4.14%) was significantly lower than that in normal and disease controls (51.9±9.73% and 47.8±5.51%, respectively) (p<0.05) (Figure 2). Moreover, a number of cell clusters (approximately 37.4%) were CK8 positive in IDD while most clusters were CK8 negative. As shown in the images, almost all cells in clusters were either CK8 positive or negative uniformly, clusters of both positive and negative cells were rarely found. No fluorescent signal could be detected in AF cells (Figure 3).
Co-expression of CK8 and CD24 by some NP cells
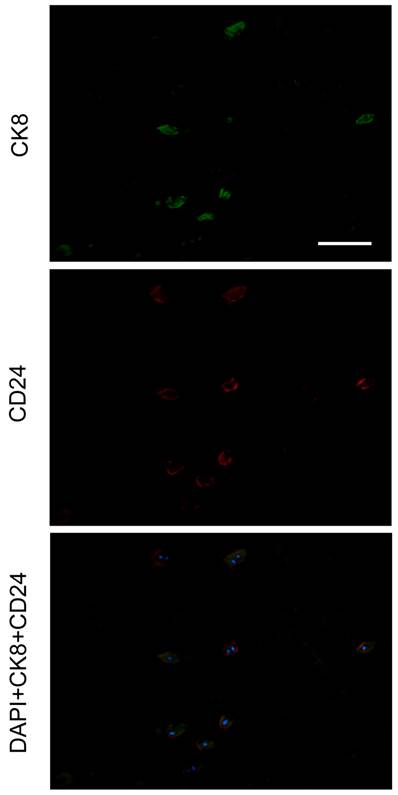
Given that CD24 is highly expressed specifically in NP [31], we performed a double-staining of CK8 and CD24. As a result, all NP cells were CD24 positive with CK8 expression in a subset of cells (Figure 4).
Human oligonucleotide primers used for real-time quantitative polymerase chain reaction analysis
| Gene | Forward primer | Reverse primer |
|---|---|---|
| CK8 | 5'-CTTCTAGGATCTCCGCCTGGTTC-3' | 5'-GACACCTTGTAGGACTTCTGGGTCA-3' |
| GAPDH | 5'-GCACCGTCAAGGCTGAGAAC-3' | 5'- TGGTGAAGACGCCAGTGGA-3' |
Western blotting results
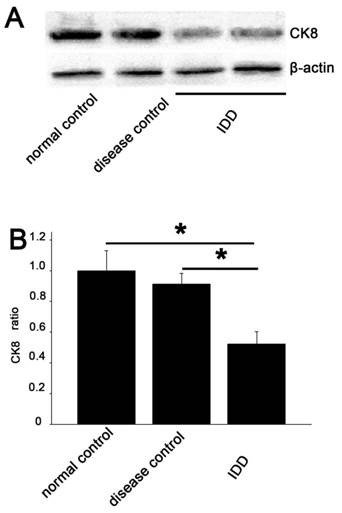
CK8 was demonstrated at a lower intensity in IDD than that in the normal and disease control. Quantitative examination revealed that the average CK8/β-actin ratio in IDD was 52.3% and 67.8% of the normal and disease control. A significant decrease in CK8 expression was demonstrated compared with the controls (p<0.05) (Figure 5).
qRT-PCR analysis
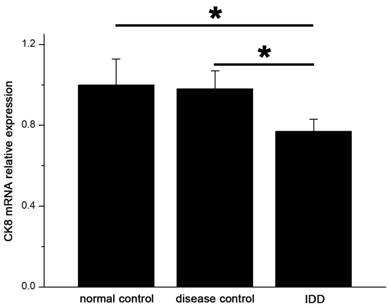
The levels of mRNA expression of CK8 in IDD were significantly lower than that in the controls. (p<0.05) (Figure 6) Relative expression of CK8 mRNA in IDD was 77.1% and 78.6% of normal and disease controls.
CK8 expression in human NP. Approximately half of NP cells were positively stained in normal and disease controls, whereas fewer cells were CK8 positive in IDD. A. specimen of a 42-year-old male from normal control, level L4/5, Grade I; B. specimen from a 17-year-old female patient with scoliosis, level L1/2, Grade II; C. Degenerative NP specimen from a 33-year-old female, level L3/4, Grade IV; D. Degenerative NP specimen from a 34-year-old male, level L4/5, Grade V. T2-weighted midsagittal MR images of each patient were showed on the offside. Scale bar=50μm.
The percentage of CK8 positive cells decreased in the NP of IDD compared with controls. Error bars represent SEM. ∗ p < 0 .05. Whole cohort of samples was used for the data analysis.
Typical staining of cell clusters in human NP. Immunofluorescence staining of a frozen specimen obtained from an IDD patient (46-year-old, male, L4/5, Grade IV) demonstrated typical cell clusters (CK8 negative and positive). CK8 positive NP cells and CK8 negative AF cells from a cadaver (45-year-old, male, L4/5, Grade I) were also provided, and nuclei were visualized with DAPI (4'-6-diamidino-2-phenylindole).Scale bar=30μm.
Double staining of CK8 and CD24 in human nucleus pulposus. Nuclei were visualized with DAPI (4'-6-diamidino-2-phenylindole). The specimen was obtained from a 42-year-old male cadaver, level L4/5, Grade I. Scale bar=100μm.
Western blotting analysis A. The degree of CK8 expression in the NP of IDD (lanes 3 and 4) was weaker than that in the controls (lanes 1 and 2), while the β-actin expression was almost the same in all samples. The 2 lines of IDD group were the outcome of two simultaneously experiments. B. Quantitative examination showed a significant decrease in CK8 expression in IDD compared with the controls (average 52.3% of the normal control and 67.8% of the disease control). Error bars represent SEM. ∗ p< 0 .05.
qRT-PCR showed significant decreased expression of CK8 in IDD at the mRNA level. Error bars represent SEM. ∗ p < 0 .05.
Discussion
Hitherto, this is the first study identifying the distribution of CK8 positive cells in human NP and their relationship with IDD based upon multiple lines of evidence to our knowledge. Moreover, the percentage of CK8 positive cell clusters was also studied. We found that the expression of CK8 could be noted thoroughly in normal NP. The percentage of CK8 positive cells decreased with disc degeneration. Meanwhile, a number of cell clusters (about 37.4%) were CK8 positive in degenerate NP. In addition, as CD24 has been described to be specific for mouse NP cells [31], we also found a high co-expression of CD24 and CK8 in a subset of human NP cells; and interestingly, almost all NP cells were CD24 positive. These findings might deepen our understanding of human NP and IDD.
First, it has been shown that CK8 was expressed in the epithelia derived notochord cells of NP [32] and was frequently used as one of the notochord cell markers to classify the origin of tissues [23,26,28,33]. The fate of notochord cells in the development of intervertebral disc has long been a controversy [34,35]. Recently, lineage-tracing experiments using Shhcre/ShhcreERT2 mouse lines and Noto-cre mouse line have shown that the notochord cells gave rise to NP in the mouse model [36,37]. However, whether all chondrocyte-like cells in human NP derived from notochord cells remains unknown. Throughout life the disc cells constantly change their phenotypes and quantity particularly in the first decade of the body [38,39]. Therefore, we suggest that CK8 positive cells might be remains of notochord cells in the development of human NP.
Second, the decreased percentage of CK8 positive cells with degeneration we found might indicate a close link between notochord remnants and the physiological function of NP. Our findings are similar to those of Weiler et al., who noted the number of CK8 positive cells was linked with distinct features of age-related disc degeneration [23]. On the other hand, we focused on the expression of CK8 in IDD excluding the contribution of age factor since the average age between IDD and normal control in this study was not statistically different. Meanwhile, given that the whole population of NP cells might decrease with degeneration [11], the down-regulation percentage of CK8 positive cells might indicate their close relationship with IDD. Indeed, notochord cells can maintain the function of NP in many aspects [19,21,40,41]. As the remnants of notochord cells, CK8 positive cells might sustain their progenitors' function in stimulating neighboring NP cells to keep vitality and normal phenotype. Actually, subpopulation of NP cells regarded as progenitor cells have been founded in the study of Sakai et al., who noted a subset of progenitor cells decreased markedly with age and degeneration of the disc. These cells were Tie2 positive and disialoganglioside 2 negative (Tie2+GD2-) in NP, suggesting the exhaustion of their capacity for regeneration [42]. However, whether these Tie2+GD2- cells are CK8 positive is unknown. Further studies are needed to address the relationship between this subpopulation and CK8 positive cells.
Third, we identified the percentage of cell clusters with CK8 positive (approximately 37.4%) for the first time. Consistent with our study, Weiler et al. showed many clustered cells with CK8 positive labeling [23]. Gilson et al. found CK8 positive cells as small isolated clusters in bovine NP and speculated cell clusters as the outcome of the proliferation of notochord cells [28]. However, the exact proportion of the CK8 positive clusters has not been stated. Meanwhile, clusters with both positive and negative cells were rarely found in the current study. This phenomenon indicates that cell clusters, which are common in IDD, might be homologous. We found a small percentage of cell clusters were CK8 positive. Several lines of evidence might support our findings. a. Using a live automated cell imaging system, Kim et al. studied notochord cells differentiation by direct observation [43]. They found notochordal cells were slower in population doubling time than chondrocyte-like cells and rarely formed clusters. b. Erwin et al. found notochord cells were essential to activate chondrocyte anabolic activity, rather than acting as a cell proliferation center [14]. c. Weiler et al. suggested notochord cells maintained a type of phagocytic activity [23]. d. Granular matrix changed around the clusters in the study of Nerlich et al., as these autophagosome lost the ability of extracellular matrix secretion [44]. e. Cytokeratins were shown to participate in autophagosome [27]. Collectively, we propose that most cell clusters are phenomenon of chondrocyte-like cells proliferation as these cells maintain a higher proliferation rate, whereas CK8 positive clusters might be autophagosomes derived from notochord cells.
Notwithstanding our study deepen the understanding of CK8 expression in IDD, we acknowledge that there are several limitations in the current study. For one, the average age of disease control, i.e., NP from patients with scoliosis, was relatively younger than that of normal and IDD groups. The variation was somewhat inevitable due to the hallmarks of adolescent scoliosis. However, we used unified MRI grading system to clarify the degeneration grades. For another, western blotting and qRT-PCR were performed from cells following in vitro culture, which might alter cell phenotype. Nevertheless, NP cells were cultured for relatively short time without cell passage to minimize the impact. Although our results provide beneficial information for CK8 expression, further studies are needed to explore the exact mechanisms between the CK8 positive cells and the neighboring NP cells.
In conclusion, this study firstly identified the distribution of CK8 positive cells in human NP. Furthermore, the percentage of cell clusters with CK8 positive (approximately 37.4%) was detected. As CK8 exerts multiple functions and is closely related to notochord cells, the CK8 positive cell subpopulation could have important implications for survival and activity of the NP. CK8 positive cells might be considered as a potential option in the development of cellular treatment strategies for NP repair.
Abbreviations
AF: annulus fibrosus; CK8: cytokeratin 8; IDD: intervertebral disc degeneration; LBP: low back pain; MRI: magnetic resonance imaging; NP: nucleus pulposus; PBS: phosphate buffered saline; qRT-PCR: quantitative real-time PCR.
Acknowledgements
The study was supported by Chinese National Natural Science Foundation Grants (No.30901509, No.81270028, No.81171747, NO. 81072116 and No.81070698).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Freemont AJ. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology (Oxford). 2009;48:5-10
2. Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88(Suppl 2):21-24
3. Biyani A, Andersson GB. Low back pain: pathophysiology and management. J Am Acad Orthop Surg. 2004;12:106-115
4. Andersson GB. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581-585
5. Uesugi K, Sekiguchi M, Kikuchi S, Konno S. The effect of repeated restraint stress in pain-related behavior induced by nucleus pulposus applied on the nerve root in rats. Eur Spine J. 2011;20:1885-1891
6. Stokes IA, Iatridis JC. Mechanical conditions that accelerate intervertebral disc degeneration: overload versus immobilization. Spine (Phila Pa 1976). 2004;29:2724-2732
7. Wang HQ, Yu XD, Liu ZH. et al. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol. 2011;225:232-242
8. Tschoeke SK, Hellmuth M, Hostmann A. et al. Apoptosis of human intervertebral discs after trauma compares to degenerated discs involving both receptor-mediated and mitochondrial-dependent pathways. J Orthop Res. 2008;26:999-1006
9. Williams FM, Bansal AT, van Meurs JB. et al. Novel genetic variants associated with lumbar disc degeneration in northern Europeans: a meta-analysis of 4600 subjects. Ann Rheum Dis. 2012
10. Aladin DM, Cheung KM, Chan D. et al. Expression of the Trp2 allele of COL9A2 is associated with alterations in the mechanical properties of human intervertebral discs. Spine (Phila Pa 1976). 2007;32:2820-2826
11. Liebscher T, Haefeli M, Wuertz K, Nerlich AG, Boos N. Age-related variation in cell density of human lumbar intervertebral disc. Spine (Phila Pa 1976). 2011;36:153-159
12. Boos N, Weissbach S, Rohrbach H. et al. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine (Phila Pa 1976). 2002;27:2631-2644
13. Roberts S, Evans H, Trivedi J, Menage J. Histology and pathology of the human intervertebral disc. J Bone Joint Surg Am. 2006;88(Suppl 2):10-14
14. Erwin WM, Inman RD. Notochord cells regulate intervertebral disc chondrocyte proteoglycan production and cell proliferation. Spine (Phila Pa 1976). 2006;31:1094-1099
15. Chen J, Yan W, Setton LA. Molecular phenotypes of notochordal cells purified from immature nucleus pulposus. Eur Spine J. 2006;15(Suppl 3):S303-S311
16. Fleming A, Keynes RJ, Tannahill D. The role of the notochord in vertebral column formation. J Anat. 2001;199:177-180
17. Pazzaglia UE, Salisbury JR, Byers PD. Development and involution of the notochord in the human spine. J R Soc Med. 1989;82:413-415
18. Trout JJ, Buckwalter JA, Moore KC. Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat Rec. 1982;204:307-314
19. Erwin WM, Ashman K, O'Donnel P, Inman RD. Nucleus pulposus notochord cells secrete connective tissue growth factor and up-regulate proteoglycan expression by intervertebral disc chondrocytes. Arthritis Rheum. 2006;54:3859-3867
20. Hunter CJ, Matyas JR, Duncan NA. The notochordal cell in the nucleus pulposus: a review in the context of tissue engineering. Tissue Eng. 2003;9:667-677
21. Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129-137
22. PEACOCK A. Observations on the postnatal structure of the intervertebral disc in man. J Anat. 1952;86:162-179
23. Weiler C, Nerlich AG, Schaaf R. et al. Immunohistochemical identification of notochordal markers in cells in the aging human lumbar intervertebral disc. Eur Spine J. 2010;19:1761-1770
24. Naka T, Iwamoto Y, Shinohara N. et al. Cytokeratin subtyping in chordomas and the fetal notochord: an immunohistochemical analysis of aberrant expression. Mod Pathol. 1997;10:545-551
25. Gotz W, Kasper M, Fischer G, Herken R. Intermediate filament typing of the human embryonic and fetal notochord. Cell Tissue Res. 1995;280:455-462
26. Stosiek P, Kasper M, Karsten U. Expression of cytokeratin and vimentin in nucleus pulposus cells. Differentiation. 1988;39:78-81
27. Galarneau L, Loranger A, Gilbert S, Marceau N. Keratins modulate hepatic cell adhesion, size and G1/S transition. Exp Cell Res. 2007;313:179-194
28. Gilson A, Dreger M, Urban JP. Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Res Ther. 2010;12:R24
29. Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976). 2001;26:1873-1878
30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408
31. Fujita N, Miyamoto T, Imai J, Hosogane N, Suzuki T. et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun. 2005;338:1890-1896
32. Han C, Zhu H. [Research progress of nucleus pulposus cells phenotypic markers]. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2011;25:867-870
33. Risbud MV, Shapiro IM. Notochordal cells in the adult intervertebral disc: new perspective on an old question. Crit Rev Eukaryot Gene Expr. 2011;21:29-41
34. Risbud MV, Schaer TP, Shapiro IM. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev Dyn. 2010;239:2141-2148
35. Henriksson HB, Brisby H. Development and Regeneration Potential of the Mammalian Intervertebral Disc. Cells Tissues Organs. 2013;197(1):1-13
36. McCann MR, Tamplin OJ, Rossant J, Séguin CA. Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5:73-82
37. Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237:3953-3958
38. Anisimov SV. [Cell therapy in treating age-related intervertebral disc pathology]. Adv Gerontol. 2012;25:105-111
39. Gantenbein-Ritter B, Chan SC. The evolutionary importance of cell ratio between notochordal and nucleus pulposus cells: an experimental 3-D co-culture study. Eur Spine J. 2012;21(Suppl 6):S819-S825
40. Erwin WM, Islam D, Inman RD, Fehlings MG, Tsui FW. Notochordal cells protect nucleus pulposus cells from degradation and apoptosis: implications for the mechanisms of intervertebral disc degeneration. Arthritis Res Ther. 2011;13:R215
41. Cappello R, Bird JL, Pfeiffer D, Bayliss MT, Dudhia J. Notochordal cell produce and assemble extracellular matrix in a distinct manner, which may be responsible for the maintenance of healthy nucleus pulposus. Spine (Phila Pa 1976). 2006;31:873-883
42. Sakai D, Nakamura Y, Nakai T. et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264
43. Kim JH, Deasy BM, Seo HY. et al. Differentiation of Intervertebral Notochordal Cells Through Live Automated Cell Imaging System In Vitro. Spine (Phila Pa 1976). 2009;23:2486-2493
44. Nerlich AG, Weiler C, Zipperer J, Narozny M, Boos N. Immunolocalization of phagocytic cells in normal and degenerated intervertebral discs. Spine (Phila Pa 1976). 2002;27:2484-2490
Author contact
![]() Corresponding author: Zhuo-Jing Luo, Department of Orthopaedics, Xijing Hospital, Fourth Military Medical University, 127 Changle Western Road Xi'an, P. R. China, 710032. E-mail: zjluoedu.cn, FAX: +86 29 84775285, Phone: +86 29 84775285.
Corresponding author: Zhuo-Jing Luo, Department of Orthopaedics, Xijing Hospital, Fourth Military Medical University, 127 Changle Western Road Xi'an, P. R. China, 710032. E-mail: zjluoedu.cn, FAX: +86 29 84775285, Phone: +86 29 84775285.

 Global reach, higher impact
Global reach, higher impact