Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(7):894-902. doi:10.7150/ijms.5556 This issue Cite
Research Paper
Evaluation of Clinical Value of Single Nucleotide Polymorphisms of Dihydropyrimidine Dehydrogenase Gene to Predict 5-Fluorouracil Toxicity in 60 Colorectal Cancer Patients in China
Department of Hematology and Oncology, China-Japan Union Hospital, Jilin University, Changchun, China, 130041.
Received 2012-11-16; Accepted 2013-5-5; Published 2013-5-20
Abstract
Dihydropyrimidine dehydrogenase (DPD) activity could be affected by single nucleotide polymorphisms (SNPs), resulting in either no effect, partial or complete loss of DPD activity. To evaluate if SNPs of DPD can be used to predict 5-FU toxicity, we evaluated five SNPs of DPD (14G1A, G1156T, G2194A, T85C and T464A) by TaqMan real time PCR in 60 colorectal cancer patients. Clinical data demonstrated that there was higher correlation between DPD activity and toxic effects of 5-FU (p<0.05). Six patients were positive for G2194A detection, which were all heterozygous. Two patients had lower DPD activities (< 3) with higher toxic effects (≥stage III) while one patient was also positive for T85C detection. Ten patients were positive for T85C detection. Two patients were homozygous with lower DPD activities and higher toxic effects. Two patients were positive for the T464A detection, which were heterozygous with lower DPD activity and higher toxic effects and also positive for T85C detection. These data clearly indicated that the T464A and homozygous of the T85C are stronger biomarkers to predict the 5-FU toxicity. Our study significantly indicated that the detection for G2194A, T85C and T464A could predict ~13% of 5-FU severe toxic side effects.
Keywords: colorectal cancer, 5-fluorouracil, Dihydropyrimidine-Dehydrogenase (DPD), single nucleotide polymorphism (SNP).
Introduction
Colorectal cancer, also called colon cancer, consists of cancerous growths in the colon, rectum and appendix. Colorectal cancer arises from adenomatous polyps in the colon. There are more than one million new cases of colorectal cancer each year in the world, and ~ half of them are fatal (1,2). In China, 130,000 - 160,000 new cases of colorectal cancer are diagnosed each year and 60,000 - 90,000 patients die. Treatment depends on the stage of the cancer (3). Surgery is still the primary treatment. Chemotherapy is also considered depending on the individual patient's staging and other underlying causes, especially whether or not it spreads to the lymph nodes (Stage III) (3). In China, treatments for colorectal cancer are surgery and chemotherapy. Most colorectal cancer patients in stages II/III are treated with surgery plus chemotherapy. Recent advances in chemotherapy treatment has led to improvement in the quality of life and survival rate in colorectal cancer patients.
5-Fuorouracil (5-FU) is a widely used anticancer drug for more than 40 years, which can cause severe toxic side effects, including death. 5-FU is also one of the most effective medicines in treating patients diagnosed with colorectal cancer (4-6). The 5-FU is a pyrimidine analog and works through non-competitive inhibition of thymidylate synthase and thus blocks the synthesis of the pyrimidine thymidine, which is a nucleotide required for DNA replication. DPD is a powerful liver enzyme to catabolize >80 % of the administered 5-FU in liver. About 8% of the population has what is termed DPD deficiency and unable to metaolize 5-FU resulting in toxic side effects because they have a genetic inability to metabolize 5-FU (7-10).
DPD gene is ~950 kb genome, which locates on chromosome 1p22, including a 3 kb coding sequence spanning in 23 exons (11,12). DPD activity is highly variable, influencing the patient's response to 5-FU, which could be due to SNP genetic polymorphism (7-9,13-18). The SNPs of DPD cause enzymatic deficiency from partial (3-5% of the population) to complete loss (0.2% of the population) of enzyme activity (7-9,13-19). More than 40 different DPD alleles have been identified (13-19). To date, the respective frequencies of different SNPs and their effects, especially in China, have not been investigated in detail. To improve the treatment for colorectal cancer, especially in China, we selected 5 SNPs of DPD, 14G1A, G1156T, G2194A, T85C and T464A. Sixty patients treated with 5-FU were evaluated to determine the correlation between the SNPs of DPD, DPD activity and the 5-FU toxic side effect.
Materials and methods
Patients and treatments
Experiments in this study were undertaken with the understanding and written consent of each subject, and such that the study conforms with the Code of Ethics of the World Medical Association. In this study, we screened 98 patients and only selected 60 patients without diarrhea before FOLFOX4 (1) scheme chemotherapy. Sixty colorectal cancer patients, 33 males and 27 females, ages between 39 and 67, were enrolled in August, 2007. They did not have any serious heart, lung, liver, kidney or brain disease, and had <4.0 x 109/L of WBC (white blood cells), <100 g/L of HB (haemoglobin), <100 x 109/L of PLT (platelet) clinically. Diagnosis of colorectal cancer for all patients was confirmed pathologically after surgery. There were 28 patients with poorly differentiated carcinoma, 16 patients with moderately differentiated carcinoma and 16 patients with well differentiated carcinoma. All patients received FOLFOX4 (1) scheme chemotherapy, which was 85 mg/m2 of oxaliplatin, 200 mg/m2 of calcium folinate and 400 mg/m2 of 5-FU intravenously plus 600 mg/m2 of 5-FU by syringe pump on day 1; 200 mg/m2 of calcium folinate and 400 mg/m2 of 5-FU intravenously plus 600 mg/m2 of 5-FU by syringe pump on day 2 - 14 for one treatment; at least two treatments/patient and 4 - 6 treatments on the average.
Determination of DPD activity
Blood samples, which used to get all DPD activities in table 4, were collected with EDTA treated blood tube at 6:00 am before breakfast two days before FOLFOX4 scheme chemotherapy, centrifuged at 3000 g for 10 min and stored at -80ºC until analysis. In this study we used ratio of dihydrouracil (UH2) to uracil (U) to represent blood DPD activity (20). The concentrations of UH2 and uracil in plasma were determined by HPLC (high-performance liquid chromatography) using C18 column (300 mm x 4.6 mm, 5μm, Dalian Elite Analytical Instruments Co., Ltd. Dalian, China) at 10ºC, 0.4 ml/min and detected with 204 nm. In this study, UH2 and uracil were extracted from 200 μl plasma. Briefly, 200 μl plasma plus 50 μl of 5-BU (5-Bromouracil) at 200μg/ml as internal control were added into a 10 ml tube and mixed by vortex for 30 min. Then, 1.5 ml of extraction buffer [n-propanol:ether (vol/vol) = 25:75] was added into the tube, mixed by vortex for 5 min, spun at 3000 g/min for 5 min and collected organic phase. This extraction procedure was repeated once and put two organic phases together. Next, the organic phase was dried at room temperature under nitrogen, re-constituted with 50 μl ddH2O, mixed by vortex for 30 s and 20 μl dichloromethane was added into the tube, mixed by vortex for 5 s and spun at 12,000 g/min for 10 min. Twenty μl supernatant were used for HPLC. The spike U/UH2 concentrations in 3% of BSA (bovine albumin) were used as a standard as following, 15.625/62.5, 31.25/125, 62.5/250, 125/500, 250/1000, 500/2000 μg/L. UH2 was detected at 204 nm. U was detected at 260 nm (21). The sensitivity was 0.01 AuFS (absorbance unit full scale). All other equipments and reagents were purchased from Fuzhou Maxim Biotechnology Technology Development Company (Fuzhou, China). Procedures were followed as described in the manual.
SNP detection
The genomic DNA from EDTA (ethylenediaminetetraacetic acid) treated peripheral blood samples were extracted using Blood Genome DNA Extraction Kit (Dalian Treasure Biotechnology; Dalian, China). Following TaqMan MGB primers and probes were purchased with plasmid Allele 1 and 2 standards as standard controls from Applied Biosystems (Foster City, CA, USA). For 14G1A, 14G1A-FP-1: TCCTCTGCAAAAATGTGAGAAGG, 14G1A-FP (or RP)-2: GCTTTTCTTTGTCAAAAGGAGACTCA, DPD-85 (T): FAM-ATGCAACTCTGTGTTCCACTTCGGC, DPD-85 (C): awakens (or TET)-CATGCAACTCTGCGTTCCACTTCG; for G2194A, DPD-2194-FP: TGAAAATGTTGATGTGTCTTGCATAG, DPD-2194-RP: CCATATGTAGTTCGCTTTGCAATC, DPD- 2194 (G): FAM-CCAATGGCGTTACAGCCACCAA, DPD- 2194 (A): awakens (or TET)-TGCCAATGGCATTACAGCCACC; for G1156T, DPD-1156-FP: GAACAAACTGCATAGCAACAATTCTC, DPD-1156-RP: TCTCTGTTCTGTTTTGTTTTAGATGGA, DPD-1156 (G): FAM-CAGAAATTCACACTTTT, DPYD-1156 (T): awakens (or TET) CAGAAATTAACACTTTTC; for T464A, DPD-464-FP: AGATATTTGTGCATGGTGATGGTAGT, DPYD-464-RP: TCCAACCTCTGATCTTTGTGTAGGT, DPD-464 (T): FAM-TGCTGCAATCCACCAA, DPD- 464 (A): awakens (or TET) TTGCTGCTATCCACCAA; for T85C: DPD-85-FP: CCTGGCTTTAAATCCTCGAACA, DPD-85-RP: GCAGTTCTTATCAGGATTTCTTTTCC, DPD-85 (T): FAM-ATGCAACTCTGTGTTCCACTTCGGC, DPD-85 (C): awakens (or TET) CATGCAACTCTGCGTTCCACTTCG. The real time PCR assays were performed in an ABI 7300 Real time PCR system using 100 ng DNA sample with TaqMan Universal PCR Master (Applied Biosystems). The conditions used were as follows: 95ºC for 10 minutes, 92ºC for 10 seconds, and 60ºC for 1 minute for 40 cycles.
Toxicity assessment
In this study, WHO toxic grading standard for chemotherapy was used (Table 1) (22). Before starting chemotherapy, patients had a very detailed physical examination and clinical lab testing. After patient received chemotherapy, blood chemistry and hematology was examined every week. Physical examination was also performed every week. Aspartate aminotransferase (AST), alanine aminotrasferase (ALT), blood urea nitrogen (BUN) and creatinine were checked to evaluate functions of liver and kidney every two weeks. We also obtained an electrocardiogram every two weeks. We carried out toxicity assessment depending on all clinical data using WHO toxic grading standard (Table 1).
Statistical analysis
Data analyses employed SPSS 13.0 software (SPSS, Inc, Chicago, IL, USA). Results herein are presented as mean values ± standard error (SEM). Chi squared test, one-way ANOVAs followed by a Tukey test and Mann Withney analysis, were employed as appropriate to determine the statistical significance of differences observed.
Results
Results of toxicity assessment of bone marrow and gastrointestinal reaction are shown in Table 2 based on criteria in Table 1. About 5-7% of patients did not have bone marrow or gastrointestinal toxicities. There were ~15% of patients with the highest grade (III-IV) of bone marrow toxicity and ~22% of patients with the highest grade (III-IV) of gastrointestinal toxicity.
Next, we examined plasma DPD activity (Fig. 1, Table 4) using liquid HPLC chromatography. It clearly showed that the patients had no or lower toxicity (toxic grades 0, I and II were defined as not severe group herein) when they had higher DPD activity in their plasma and higher toxicity when they had lower DPD activity in their plasma (p< 0.001 by one-way ANOVAs and Mann Withney U = 18.50, two-tailed p<0.0001 for bone marrow inhibition; p< 0.001 by one-way ANOVAs and Mann Withney U = 149.0, two-tailed p<0.0115 for gastrointestinal reaction).
To understand the correlation between DPD activity, toxicity of 5-FU and SNPs of DPD, we detected 5 SNPs of DPD, 14G1A, G1156T, G2194A, T85C and T464A, using genomic DNAs from all 60 patients. Characteristics of each SNP are shown in Table 2. Results of SNPs by real time PCR are shown in Table 3. All 60 patients were negative for 14G1A and G1156T detection. Six patients (10%) were heterozygous changes for G2194A. Ten patients (~16.7%) were positive for T85C, whereas 8 patients (~13.3%) heterozygous changes and 2 patients (~3.3%) were homozygous changes. For T464A, 2 patients (~3.3%) were heterozygous changes.
Data in Table 4 demonstrated that there was no gender and age correlation with SNPs of DPD, G2194A, T85C and T464A. Six positive patients for G2194A (Table 3 and 4) were all heterozygous. Four patients had higher DPD activities (> 3) with corresponding lower toxic side effects (≤ stage II), and two patients had lower DPD activities (< 3) with corresponding higher toxic side effects (≥stage III), and one patient was also positive for T85C detection. Ten patients (Table 4) were positive for T85C detection. In eight patients who had heterozygous changes, three patients had lower DPD activities (< 3) with corresponding higher toxic side effects (≥stage III), and five patients had higher DPD activities (> 3) with stage I to IV toxic side effects. Two patients for T85C detection (Table 4) were homozygous with lower DPD activities (< 3) and higher toxic side effects (≥stage III). Only two patients were positive to the T464A detection, which were heterozygous with lower DPD activity and higher toxic side effects, especially bone marrow inhibition. These two patients were also positive for T85C detection.
Statistical analyses demonstrated that the overall DPD activity was significant lower in SNP positive patients. In this study, the patients with two SNP positive or homozygous changes had markedly lower DPD activity than that of SNP negative patients (p< 0.001 - 0.007 by one-way ANOVAs)(Fig. 2A-2D). Fig. 2E also demonstrated that the DPD activity was significantly lower in two SNP positive or homozygous patients than that of one SNP positive patients (p< 0.004 by one-way ANOVAs). Fig. 3 illustrates the comparison of toxic grade differences between SNP positive and SNP negative patients. Fig. 3 indicates that the toxic grades of bone marrow inhibition or gastrointestinal reaction from SNP positive patients was significantly higher than that of SNP negative patients (p< 0.0005 or 0.0072 by Mann Withney analysis). Table 5 shows that the SNP positive patients with severe toxic grades of bone marrow inhibition or gastrointestinal reaction were significantly higher than that of the SNP negative patients with severe toxic grades of bone marrow inhibition or gastrointestinal reaction (p< 0.0001 or 0.0019 by Chi-square analysis) while no-severe toxic grades were not significantly different.
These data indicated that the DPD activity was significantly lower in the patient with severe toxic grade of bone marrow inhibition or gastrointestinal reaction as well as in the SNP positive patients. Also, the toxic grade was higher in the SNP positive patients. The T464A and homozygous of T85C have been shown to be significantly correlated to the lower DPD activity and higher 5-FU induced bone marrow inhibition. Two positive SNPs, either G2194A, T85C or T464A, in one patient were significantly correlated with severe 5-FU toxic side effects, especially bone marrow inhibition.
Criteria of bone marrow toxicity and gastrointestinal toxicity used in current study [29].
| Grades | |||||
|---|---|---|---|---|---|
| 0 | I | II | III | IV | |
| Bone marrow inhibition | |||||
| HB (g/L) | ≥110 | 95-109 | 80-94 | 65-79 | <65 |
| WBC (x109/L) | ≥4.0 | 3.0-3.9 | 2.0-2.9 | 1.0-1.9 | 1.0 |
| PLT (x109/L) | >100 | 75-99 | 50-74 | 25-49 | <25 |
| Gastrointestinal reaction | |||||
| AST | ≤1.25xN | 1.26-2.5xN | 2.6-5xN | 5.1-10.0xN | >10xN |
| Nausea and vomiting | None | Nausea | Transient vomiting | Vomiting requiring therapy | Intractable vomiting |
Note: HB, hemoglobin; WBC. White blood cells; PLT, platelets; N, normal (5 - 40 IU/L).
Characteristics of 5 SNPs of DPD used in this study in the literature.
| Mutation | Exon | Protein | Frequency (%) | Consequence | International code | Reference |
|---|---|---|---|---|---|---|
| 14G1A | 14 | Del(exon 14) | 1.5 | No expression | rs1801267 | 15,28.29 |
| G1156T | 11 | E386Ter | NA | Nonsense, truncated protein, no activity | rs1333728 | 29,31 |
| G2194A | 18 | V732I | 5 | Expression↓ | rs1801160 | 29 |
| T85C | 2 | C29R | 29 | Unclear | rs1801265 | 32,33 |
| T464A | 5 | stop codon in the protein | 0.2 | Expression↓ | rs666523971 | 34 |
Summary of 5 SNPs of DPD in 60 patients with colorectal cancer.
| Wild type | Heterozygous (genotype frequency) | Homozygous (genotype frequency) | % of SNP | |
|---|---|---|---|---|
| 14G1A | 60 | 0 | 0 | 0 |
| G1156T | 60 | 0 | 0 | 0 |
| G2194A | 54 | 6 (10%) | 0 | 10 |
| T85C | 50 | 8 (13.3%) | 2 (3.3%) | 16.7 |
| T464A | 58 | 2 (3.3%) | 0 | 3.3 |
Summary of all 60 cases.
| Case | Gender | Age | Diarrhea | BMI | GIR | DPD activity | G2194A | T85C | T464A | |
|---|---|---|---|---|---|---|---|---|---|---|
| Before | After | |||||||||
| 1 | M | 68 | No | No | IV | IV | 2.57 | C/C | T/A | |
| 2 | M | 63 | No | No | IV | IV | 2.02 | C/C | ||
| 3 | F | 47 | No | No | IV | III | 2.19 | G/A | T/C | |
| 4 | F | 46 | No | No | IV | II | 2.23 | T/C | T/A | |
| 5 | F | 42 | No | No | I | I | 3.92 | T/C | ||
| 6 | F | 40 | No | No | II | III | 3.65 | T/C | ||
| 7 | M | 67 | No | No | II | III | 3.87 | T/C | ||
| 8 | M | 50 | No | No | II | II | 3.64 | T/C | ||
| 9 | F | 50 | No | No | III | III | 3.65 | T/C | ||
| 10 | M | 67 | No | No | III | IV | 2.07 | T/C | ||
| 13 | M | 59 | No | No | I | I | 4.48 | G/A | ||
| 11 | F | 40 | No | No | II | II | 3.23 | G/A | ||
| 12 | M | 61 | No | No | II | II | 3.46 | G/A | ||
| 14 | M | 58 | No | I | III | I | 3.30 | G/A | ||
| 15 | F | 65 | No | No | III | III | 2.60 | G/A | ||
| 24 | M | 49 | No | No | 0 | I | 5.23 | |||
| 28 | M | 47 | No | No | 0 | 0 | 5.03 | |||
| 29 | M | 54 | No | No | 0 | I | 4.23 | |||
| 16 | F | 52 | No | No | I | II | 4.78 | |||
| 20 | M | 55 | No | No | I | III | 5.80 | |||
| 21 | F | 44 | No | No | I | 0 | 5.47 | |||
| 22 | M | 50 | No | No | I | I | 3.96 | |||
| 23 | F | 41 | No | No | I | 0 | 3.98 | |||
| 25 | F | 50 | No | No | I | I | 3.98 | |||
| 26 | F | 43 | No | No | I | II | 4.79 | |||
| 30 | M | 64 | No | No | I | III | 5.67 | |||
| 31 | F | 48 | No | No | I | I | 5.16 | |||
| 32 | M | 63 | No | No | I | II | 4.01 | |||
| 17 | F | 52 | No | No | II | 0 | 4.12 | |||
| 18 | F | 52 | No | I | II | 0 | 2.85 | |||
| 19 | M | 39 | No | No | II | II | 4.63 | |||
| 27 | F | 67 | No | No | II | I | 4.84 | |||
| 33 | M | 50 | No | No | II | II | 5.02 | |||
| 34 | M | 49 | No | No | II | II | 3.78 | |||
| 35 | M | 60 | No | No | II | II | 4.28 | |||
| 36 | M | 39 | No | No | II | II | 4.03 | |||
| 37 | F | 51 | No | No | II | II | 2.95 | |||
| 38 | M | 49 | No | No | II | II | 4.21 | |||
| 39 | M | 67 | No | No | II | II | 4.49 | |||
| 40 | M | 59 | No | No | II | II | 3.72 | |||
| 41 | F | 56 | No | No | II | II | 4.42 | |||
| 43 | M | 48 | No | No | II | II | 3.91 | |||
| 44 | M | 63 | No | No | II | II | 5.62 | |||
| 45 | M | 58 | No | No | II | II | 4.21 | |||
| 46 | M | 51 | No | I | II | II | 3.69 | |||
| 47 | F | 48 | No | No | II | II | 4.68 | |||
| 48 | F | 56 | No | No | II | II | 5.22 | |||
| 49 | F | 52 | No | No | II | II | 4.58 | |||
| 50 | F | 58 | No | No | II | II | 4.28 | |||
| 51 | F | 62 | No | No | II | II | 4.30 | |||
| 52 | F | 57 | No | No | II | II | 4.27 | |||
| 53 | M | 52 | No | I | II | II | 4.33 | |||
| 54 | F | 59 | No | No | II | II | 4.62 | |||
| 55 | M | 61 | No | No | II | II | 4.88 | |||
| 56 | M | 60 | No | No | II | II | 4.57 | |||
| 57 | F | 58 | No | No | II | II | 3.60 | |||
| 58 | M | 51 | No | No | II | II | 3.99 | |||
| 59 | M | 51 | No | No | II | IV | 2.52 | |||
| 60 | F | 58 | No | No | II | II | 3.85 | |||
| 42 | F | 45 | No | No | III | II | 3.49 | |||
Note: M, male; F, female; BMI, bone marrow inhibition; GIR, gastrointenstinal reaction. Before: before FOLFOX4 chemotherapy; Aftr: after FOLFOX4 chemotherapy.
Comparisons between numbers of SNP and toxic grades.
| Grade of bone marrow toxicity | ||
| I - II | III - IV | |
| No. of patients had one SNP | 7 | 5 |
| No. of patients had two SNPs | 0 | 3 |
| No. of patients had no SNP | 44 | 1 |
| Chi-square analysis between +SNP and -SNP groups | ||
| Chi-square | 2.205 | 14.38 |
| Odds ratio | 0.4773 | 24.00 |
| p | 0.4773 | 0.0001 |
| Grade of gastrointestinal toxicity | ||
| I - II | III - IV | |
| No. of patients had one SNP | 6 | 6 |
| No. of patients had two SNPs | 1 | 2 |
| No. of patients had no SNP | 42 | 3 |
| Chi-square analysis between +SNP and -SNP groups | ||
| Chi-square | 1.922 | 9.669 |
| Odds ratio | 0.500 | 8.00 |
| p | 0.1656 | 0.0019 |
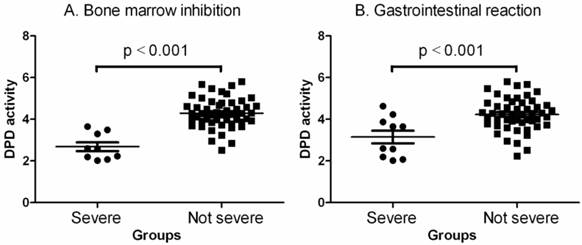
Comparisons of DPD activities between no severe (grades 0, I and II) and severe (grades III and IV) toxic groups. A. Bone marrow inhibition. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 4.292, lower 95% CI = 4.092, upper 95% CI = 4.492, p<0.001 by one-way ANOVAs and Mann Withney U = 18.50, two-tailed p<0.0001). B. Gastrointestinal reaction. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 4.050, lower 95% CI = 3.816, upper 95% CI = 4.284, p<0.001 by one-way ANOVAs and Mann Withney U = 149.0, two-tailed p<0.0115).
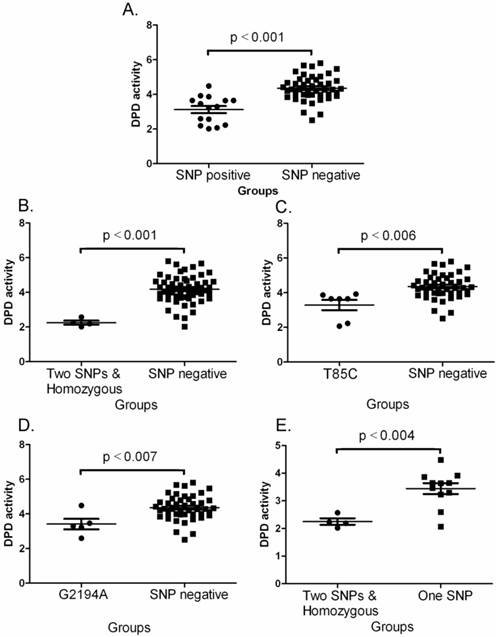
Comparisons of DPD activities between various groups. A. Between SNP negative and SNP positive. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 3.125, lower 95% CI = 2.692, upper 95% CI = 3.559, p<0.001 by one-way ANOVAs and Mann Withney U = 70.0, two-tailed p<0.0001). B. Between SNP negative and two SNPs plus homozygous. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 4.177, lower 95% CI = 3.966, upper 95% CI = 4.388, p<0.001 by one-way ANOVAs and Mann Withney U = 4.0, two-tailed p<0.0014). C. Between SNP negative and T85C positive. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 3.467, lower 95% CI = 2.737, upper 95% CI = 4.196, p<0.006 by one-way ANOVAs and Mann Withney U = 34.0, two-tailed p<0.0033). D. Between SNP negative and G2194A positive. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 3.414, lower 95% CI = 2.570, upper 95% CI = 4.258, p<0.007 by one-way ANOVAs and Mann Withney U = 36.0, two-tailed p<0.014). E. Between one SNP and two SNPa plus homozygous. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 2.253, lower 95% CI = 1.886, upper 95% CI = 2.619, p<0.004 by one-way ANOVAs and Mann Withney U = 3.0, two-tailed p<0.0157).
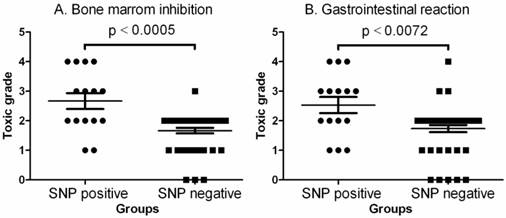
Comparison of toxic grades between SNP positive and SNP negative patients. A. Comparison of toxic grade of bone marrow inhibition. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 2.667, lower 95% CI = 2.087, upper 95% CI = 3.246, two-tailed p<0.0005 by Mann Withney analysis and Mann Withney U = 158.5). B. Comparison of toxic grade of gastrointestinal reaction. The data shown are mean values ± SD. Differences between two groups are significant (Mean = 2.533, lower 95% CI = 1.946, upper 95% CI = 3.120, two-tailed p<0.0005 by Mann Withney analysis and Mann Withney U = 196.5).
Discussion
Recently, many studies demonstrated that DPD could not efficiently metabolize 5-FU due to DPD genomic variation (13-16), such as exon skipping (23), deletion (24) and missense mutation (25-27). The SNP of DPD is one of the major reasons for DPD deficiency. During our clinical practice, it was recognized that 83.3% of 5-FU treated patients had severe side effects resulting in higher risk for the patients and less or no effects of chemotherapy. To overcome this difficulty and predict toxicity before treatment, clinicians have tried to determine if the SNP of DPD could be used as a biomarker to improve clinical treatment. We, however, still understand little about these SNPs, especially in different populations such as the Chinese population.
It was known that ~50% of patients, who had severe toxicity of 5-FU, were genotypically heterozygous or homozygous for known SNPs in the DPD gene (14,28,29). In the present study, we assessed 5 SNPs of DPD gene for 60 colorectal cancer patients to evaluate if these SNPs could be used to predict the possible toxicity of 5-FU before treatment.
Our results demonstrated that lower activity of DPD was significantly correlated to higher 5-FU toxicity, and higher activity of DPD was also significantly correlated to lower 5-FU toxicity (Table 4, Fig. 1), after we screened 5 known SNPs of DPD, 14G1A, G1156T, G2194A, T85C and T464A, in 60 colorectal cancer patients. Results indicated that the SNPs of 14G1A and G2194A were not common SNPs to affect DPD activity in Chinese, although the 14G1A seem to be very common for other populations (Table 2 and 3) (15,28,30). The frequency of T464A was 3.3%, 10% for G2194A and 16.7% for T85C. Interestingly, the DPD activity was statistically lower in the SNP positive patients while the toxic grade of bone marrow inhibition or gastrointestinal reaction was higher in the SNP positive patients (Tables 4 and 5, Figs. 2 and 3). Especially, the T464A, homozygous of the T85C or any combination between T464A, G2194 and T85C were highly correlated with the lower DPD activity and the bone marrow inhibition than that for gastrointestinal reaction (Tables 3 and 4, Fig. 2).
In summary, we have shown that the SNPs of 14G1A and G2194A were not common in Chinese and indicated that these two SNPs could have differential race expression. Furthermore, we have observed that the SNPs of T464A, homozygous of the T85C or combination of T464A, G2194 and T85C can be used as stronger biomarkers to predict 5-FU toxicity for Chinese population, especially for the bone marrow inhibition (~5%). To further confirm the clinical value of these three SNPs, we need to screen more patients in order to use these SNPs as a clinical marker to prevent the risk of the 5-FU toxicity before treatment.
Acknowledgements
This work was supported by grants from the Jilin scientific developing projects (No: 20050405-4).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Parkin DM, Bray F, Ferlay J. et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108
2. SEER Stat Fact Sheet; colon and rectum. National Cancer Institute. http://seer.cancer.gov/statfacts/html/colorect.html
3. Ades S. Adjuvant chemotherapy for colon cancer in the elderly: moving from evidence to practice. Oncology. 2009;23:162-167
4. Tuchman M, Stoeckeler JS, Kiang DT. et al. Familial pyrimidinemia and pyrimidinuria associated with severe fluorouracil toxicity. N Engl J Med. 1985;313:245-249
5. Diasio RB, Beavers TL, Carpenter JT. Familial deficiency of dihydropyrimidine dehydrogenase. Biochemical basis for familial pyrimidinemia and severe 5-fluorouracil-induced toxicity. J Clin Invest. 1988;81:47-51
6. Diasio RB, Johnson MR. Dihydropyrimidine dehydrogenase: its role in 5-fluorouracil clinical toxicity and tumor resistance. Clin Cancer Res. 1999;5:2672-2673
7. Pinedo HM, Peters GF. Fluorouracil: biochemistry and pharmacology. J Clin Oncol. 1988;6:1653-1664
8. Etienne MC, Lagrange JL, Dassonville O. et al. Population study of dihydropyrimidine dehydrogenase in cancer patients. J Clin Oncol. 1994;12:2248-2253
9. Lu Z, Zhang R, Diasio RB. Dihydropyrimidine dehydrogenase activity in human peripheral blood mononuclear cells and liver: population characteristics, newly identified deficient patients, and clinical implication in 5-fluorouracil chemotherapy. Cancer Res. 1993;53:5433-5438
10. van Kuilenburg AB, De Abreu RA, van Gennip AH. Pharmacogenetic and clinical aspects of dihydropyrimidine dehydrogenase deficiency. Ann Clin Biochem. 2003;40:41-45
11. Takai S, Fernandez-Salguero P, Kimura S. et al. Assignment of the human dihydropyrimidine dehydrogenase gene (DPYD) to chromosome region 1p22 by fluorescence in situ hybridization. Genomics. 1994;24:613-614
12. Wei X, Elizondo G, Sapone A. et al. Characterization of the human dihydropyrimidine dehydrogenase gene. Genomics. 1998;51:391-400
13. Van Kuilenburg AB, Vreken P, Abeling NG. et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum Genet. 1999;104:1-9
14. van Kuilenburg AB, Haasjes J, Richel DJ. et al. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5-fluorouracil-associated toxicity: identification of new mutations in the DPD gene. Clin Cancer Res. 2000;6:4705-4712
15. van Kuilenburg AB, Muller EW, Haasjes J. et al. Lethal outcome of a patient with a complete dihydropyrimidine dehydrogenase (DPD) deficiency after administration of 5-fluorouracil: frequency of the common IVS14+1G>A mutation causing DPD deficiency. Clin Cancer Res. 2001;7:1149-1153
16. van Kuilenburg AB, Dobritzsch D, Meinsma R. et al. Novel disease-causing mutations in the dihydropyrimidine dehydrogenase gene interpreted by analysis of the three-dimensional protein structure. Biochem J. 2002;364:157-163
17. van Kuilenburg AB, Häusler P, Schalhorn A. et al. Evaluation of 5-fluorouracil pharmacokinetics in cancer patients with a c.1905+1G>A mutation in DPYD by means of a Bayesian limited sampling strategy. Clin Pharmacokinet. 2012;51:163-174
18. Zhang XP, Bai ZB, Chen BA. et al. Polymorphisms of dihydropyrimidine dehydrogenase gene and clinical outcomes of gastric cancer patients treated with fluorouracil-based adjuvant chemotherapy in Chinese population. Chin Med J (Engl). 2012;125:741-746
19. He YF, Wei W, _ Zhang X. et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in Chinese cancer patients. J Clin Pharm Ther. 2008;33:307-314
20. Mercier C, Yang C, Ciccolini J. et al. Determination of uracil/UH2 ratio as a potential surrogate for DPD status in cancer patients presenting with severe toxicities during fluoropyrimidine treatment. J Clin Oncol ASCO Annual Meeting Proceedings Part 1. 2006;24:2020
21. Li S, Lai H, Li Y, Zhang L, Lin T, He Y. Determination of endogenous uracil and dihydrouracil in blood of cancer patients. Chinese Journal of Clinical Pharmacy. 2005;14:345-348
22. World Health Orgnization. Table 1 Recommendations for grading of acute and subacute toxic effects.; In WHO Handbook for Reporting Results of Cancer Treatment. Switzerland: WHO offset publication. 1979:15-16
23. Meinsma R, Fernandez-Salguero P, Van Kuilenburg ABp. et al. Human polymorphism in drug metabolism: mutation in the dihydropyrimidine dehydrogenase gene results in exon skipping and thymine uracilurea. DNA Cell Biol. 1995;14:1-6
24. P. Vreken P, Van Kuilenburg ABP, Meinsma R, et al. Identification of a four-base deletion (delTCAT296-299) in the dihydropyrimidine dehydrogenase gene with variable clinical expression. Hum. Genet. 1997;100:263-265
25. Vreken P, Van Kuilenburg ABP, Meinsma R. et al. Dihydropyrimidine dehydrogenase (DPD) deficiency: identification and expression of missense mutations C29R, R886H and R235W. Hum. Genet. 1997;101:333- 338
26. Van Kuilenburg ABP, Vreken P, Riva D. et al. Clinical and biochemical abnormalities in a patient with dihydropyrimidine dehydrogenase deficiency due to homozygosity for the C29R mutation. J. Inherit. Metab. Dis. 1999;22:191-192
27. Van Kuilenburg ABP, Vreken P, Abeling NG. et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Hum. Genet. 1999;104:1-9
28. McLeod HL, Collie-Duguid ES, Vreken P. et al. Nomenclature for human DPYD alleles. Pharmacogenetics. 1998;8:455-459
29. Van Kuilenburg AB, Meinsma R, Zoetekouw L. et al. Increased risk of grade IV neutropenia after administration of 5-fluorouracil due to a dihydropyrimidine dehydrogenase deficiency: high prevalence of the IVS14+1g>a mutation. Int J Cancer. 2002;101:253-258
30. Robert J, Morvan VL, Smith D. et al. Predicting drug response and toxicity based on gene polymorphisms. Crit Rev Oncol Hematol. 2005;54:171-196
31. Kouwaki M, Hamajima N, Sumi S. et al. Identification of novel mutations in the dihydropyrimidine dehydrogenase gene in a Japanese patient with 5-fluorouracil toxicity. Clin Cancer Res. 1998;4:2999-3004
32. Vreken P, Van Kuilenburg AB, Meinsma R. et al. Dihydropyrimidine dehydrogenase (DPD) deficiency: identification and expression of missense mutations C29R, R886H and R235W. Hum Genet. 1997;101:333-338
33. Vreken P, van Kuilenburg AB, Meinsma R. et al. Dihydropyrimidine dehydrogenase deficiency: a novel mutation and expression of missense mutations in E. coli. J Inherit Metab Dis. 1998;21:276-279
34. Morel A, Boisdron-Celle M, Fey L. et al. Identification of a novel mutation in the dihydropyrimidine dehydrogenase gene in a patient with a lethal outcome following 5-fluorouracil administration and the determination of its frequency in a population of 500 patients with colorectal carcinoma. Clin Biochem. 2007;40:11-17
Author contact
![]() Corresponding author: Dr. Zhenxia Lu, Department of Hematology and Oncology, China-Japan union Hospital, Jilin University, Changchun, China, 130031. Tel: 86-431-84995812 Email: luzxedu.cn.
Corresponding author: Dr. Zhenxia Lu, Department of Hematology and Oncology, China-Japan union Hospital, Jilin University, Changchun, China, 130031. Tel: 86-431-84995812 Email: luzxedu.cn.

 Global reach, higher impact
Global reach, higher impact