Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(6):719-729. doi:10.7150/ijms.5037 This issue Cite
Research Paper
Inhibition of Dengue Virus Entry into Target Cells Using Synthetic Antiviral Peptides
1. Department of Medical Microbiology, Faculty of Medicine, University of Malaya, Lembah Pantai, 50603 Kuala Lumpur, Malaysia;
2. Department of Medical Microbiology, International Medical School, Management & Science University, Shah Alam, 40100, Selangor, Malaysia;
3. Virtual Reality Centre/ Centre for Biomedical Technology Intergration Pte Ltd, University of Malaya, 50603 Kuala Lumpur, Malaysia;
4. Institute of Medical Molecular Biotechnology, Faculty of Medicine, Universiti Teknologi Mara, Selangor, Malaysia;
5. Department of Trauma and Emergency Medicine, University Malaya Medical Centre, Kuala Lumpur, Malaysia.
Received 2012-8-14; Accepted 2013-3-7; Published 2013-4-16
Abstract
Despite the importance of DENV as a human pathogen, there is no specific treatment or protective vaccine. Successful entry into the host cells is necessary for establishing the infection. Recently, the virus entry step has become an attractive therapeutic strategy because it represents a barrier to suppress the onset of the infection. Four putative antiviral peptides were designed to target domain III of DENV-2 E protein using BioMoDroid algorithm. Two peptides showed significant inhibition of DENV when simultaneously incubated as shown by plaque formation assay, RT-qPCR, and Western blot analysis. Both DET4 and DET2 showed significant inhibition of virus entry (84.6% and 40.6% respectively) using micromolar concentrations. Furthermore, the TEM images showed that the inhibitory peptides caused structural abnormalities and alteration of the arrangement of the viral E protein, which interferes with virus binding and entry. Inhibition of DENV entry during the initial stages of infection can potentially reduce the viremia in infected humans resulting in prevention of the progression of dengue fever to the severe life-threatening infection, reduce the infected vector numbers, and thus break the transmission cycle. Moreover these peptides though designed against the conserved region in DENV-2 would have the potential to be active against all the serotypes of dengue and might be considered as Hits to begin designing and developing of more potent analogous peptides that could constitute as promising therapeutic agents for attenuating dengue infection.
Keywords: Dengue virus, Antiviral peptides, Inhibitory Peptides, Viral entry, Envelope protein, Domain III
Introduction
DENV entry into the host cells is critical to establish the infection. It is mediated by viral E protein in both attachments to cellular surface receptors and internalization into the host cell [1-3]. The E protein plays crucial functional roles in viral infection, through virus attachment to cell-surface receptors and mediating viral-cell membrane fusion. It is the main target for protective antibodies against dengue [1]. It consists of three domains namely a central structural domain (domain I), a dimerization domain (domain II), and receptor-binding domain (domain III) [4].
Antimicrobial peptides are an essential part of natural host defense mechanism. In some cases, they have direct antimicrobial effect against parasites, fungi, bacteria, and viruses. They can damage or disrupt the cell membrane of invading pathogens or affect targets involved in the organization of the innate immune and inflammatory responses [5]. There are four main structural classes of antimicrobial peptides including β-sheet structure, amphipathic α-helices structure, loop structure, and extended structure [6]. It is difficult to predict the activity of the peptides based on secondary structures and other physical properties. For example, some of α-helical peptides have shown low antiviral activity against HSV such as cathelicidin LL-37 [7], while other α-helical peptides such as magainins have shown some anti-HSV activity [8]. On the other hand, ß-sheet peptides such as tachyplesin, defensins, and protegrins have high antiviral activity toward HSV [9]. Furthermore, there is variation in antiviral activity within the same peptide subclasses. For example, variation from highly active to inactive against HSV infections was observed in protegrin analogues that lack one or both disulphide bounds [10]. The advanced progress in the structural characteristics of DENV E protein has enabled the use of physico-chemical algorithms in combination with known structural information to identify potential peptide inhibitors of DENV entry. These synthetic peptides could specifically block viral entry and fusion with the cell membrane by targeting different structures of the viral E protein and thus could help towards the development of optimized peptide drugs [11, 12].
This study aimed to investigate the effect of targeting the DENV E protein by inhibitor peptides to block DENV entry into target cells, which eventually leads to inhibition of viral infection. This was performed by designing four antiviral peptides to target domain III of DENV-2 E protein based on the calculation of the sum of hydrophobic and charge compatibility index using the BioMoDroid algorithm. Then the antiviral activity of these peptides was evaluated in vitro experimentally.
Methods
Cells and virus
LLC-MK2 cells with passage number of less than 50 were maintained in DMEM media supplemented with 10% FBS at 37°C in 5% CO2. LLC-MK2 cells were used to study the peptides cytotoxicity and antiviral activity. DENV-2 (strain New Guinea C) was propagated in C6/36 cells (cat #: CRL-1660, ATCC, USA) and stored at −80°C till use as described previously [13]. Virus was titrated using the plaque assay on porcine kidney cells (PS clone D) as described previously [14].
Design and synthesis of antiviral peptides
The polypeptide sequence of domain III of DENV E protein was obtained from Protein Data Bank (PDB ID: 2JSF). A short sequence (residues 380-IGVEPGQLKL-389) in the lateral loop on domain III of DENV-2 E protein which has been implicated in receptor-binding of DENV-2 was selected as a target for peptides binding based on previous literature [15, 16]. Novel peptide design was done by using the BioMoDroid algorithm developed in C++ programming language at the Virtual Reality Centre/Centre for Biomedical Technology Integration Pte Ltd, University of Malaya. A HPC system consisting of two nodes with each node having an Intel Quad Core 2.40 GHz processor, 8 Gigabytes of Random Access Memory and 5 Terabytes of hard disk storage (from Hewlett Packard), was used to process the calculations. The algorithm calculated a combined score for each sequence based on iterative pairwise scores comparing each amino acid on the receptor/interface against its counterpart on the ligand. The receptors were auto generated iteratively with all the possible amino acid sequences for a fixed length of 10 amino acids. This involved 2010 combinations and in order to accelerate the result acquisition, each node on the HPC was programmed to process multiple sequences simultaneously using a multiple distributed child process method. The pairwise score was calculated by iteratively varying the ratio of values of the hydrophobicity index and charge compatibility index derived from indices [17]. The results for each peptide-receptor complex were then tabulated and organized in a database management system MySQL Ver 14.14 Distrib 5.1.46. Top antiviral peptide sequences were selected based on the final score which we termed as the BioMoDroid Score. Selected peptides were then compared with the protein database and no matching or similarities were found. Finally, peptides were custom chemically synthesized using solid-phase chemistry by JPT Peptide Technologies GmbH, Berlin, Germany. Peptides were purified (>90%) by reverse-phase high-performance liquid chromatography. The purity was confirmed by mass spectrometry.
Cytotoxicity
To determine the Toxicity of peptides, cells were exposed to increasing concentrations (1 to 500 µM final concentration) of the peptides for 24, 48, and 72 hours. Cytotoxicity was measured by estimating the number of viable cells compared to untreated cells using CellTiter 96® Aqueous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI) in accordance to the manufacturer's protocol.
Plaque formation assay
Plaque formation assay was performed essentially as previously described [18] using LLC-MK2 cells at a density of 3×105 cells/well in 6-wells plate and approximately 200 PFU of the DENV-2. Scoring was done by counting the plaques in each well and normalized to untreated controls to calculate the inhibition percentage.
Dengue virus entry and infection
LLC-MK2 cells were incubated with a mixture of desired peptides dilution and MOI of 2 of DENV-2 in 6-wells plate. Plates were incubated for one hour at room temperature and then for three hours at 37 °C with 5% CO2 and rocked every 15 minutes. The unattached viruses were then washed away with PBS three times. Cells were re-incubated for 24 hours at 37 °C with 5% CO2. The viral load was then evaluated by RT-qPCR and Western blot analysis.
For the post-infection antiviral activity of DET4 peptide evaluation, the peptide was added directly to the cells 24 hours post-infection. Cells were incubated for one hour at room temperature and then three hours at 37 °C with 5% CO2 and rocked every 15 minutes. The supernatant was then washed away with two milliliters of sterile PBS three times. After that, cells were re-incubated for 24 hours before viral load evaluation was carried out using RT-qPCR.
Viral RNA quantification
Total cellular RNA was extracted using RNeasy® Plus mini Kit (QIAGEN, Germany) according to the manufacturer's specifications. The purity and concentration of extracted RNA samples were checked using Implen NanoPhotometerTM. One-step RT-qPCR was carried out in CFX96™ Real-Time PCR Detection System using the iScriptTM One-Step RT-PCR Kit with SYBR® Green (Bio-Rad, USA). The primers sequence, reaction condition, and thermal cycling profile were performed using the method of Kong et al [19]. The specificity of the amplicon was verified by melt curve analysis. Viral RNA load in peptide treated cells was compared to the untreated controls and was normalized to the reference gene (β-actin). Data is expressed as relative fold expression to untreated controls, which was defined as 1.0 fold (100%). Triplicate reactions were carried out for each sample, and no template control was included as a negative control.
Western blot analysis
Lysates were prepared for immunoblotting against dengue viral antigen using ice-cold lysis buffer. The amount of protein in cell lysates was quantified to ensure equal loading (20 μg ) in western blot gels using 2-D Quant Kit (GE Healthcare Bio-Sciences, USA) according to the manufacturer's specifications. All gels were prepared freshly and two separate gels, resolving and stacking gel, were used. The experiment includes a well for the protein marker ladder (Precision Plus Protein Kaleidoscope Standard with range of 7.6-216 kDa, Bio-Rad Laboratories, USA). The separated proteins were transferred onto nitrocellulose membrane and then blocked with blocking buffer. Membranes were incubated overnight with dengue infected human serum containing anti-dengue IgG at 1:500. After washing three times, the membranes were incubated with Polyclonal Rabbit Anti-Human IgG conjugated with horse-radish peroxidase (Dako, Denmark) at 1:1,000 for two hours. Horseradish peroxidase substrate was added to develop the color. The membrane was then scanned to get a digital image and the relative levels of DENV-2 E protein in peptide treated cells were determined by quantitative western blotting using image analysis (Image J). Results were compared to the untreated controls, which was defined as one fold and then were normalized against β-actin.
Transmission electron microscopy
Purified DENV-2 was mixed with the peptides at a concentration of 500 µM. The complex was incubated for one hour at 37 °C. The copper grids were suspended in the virus-peptides mixture for 3-5 minutes and then dried on filter paper. The grids were then re-suspended with negative stain Phosphotunstic Acid (PTA) for 2-4 minutes and then dried on filter paper. Images of the virus were taken with Leo Libra 120 TEM (Carl Zeiss Microscopy, Germany) at The Electron Microscopy Unit- Faculty of Medicine University of Malaya with actual magnification of 100,000 X.
Statistical analysis
All assays, including cytotoxicity assay, quantification of viral RAN by RT-qPCR, plaque assay, and Western plot analysis were done in triplicates. All statistical analyses were performed using GraphPad Prism version 5.01 (GraphPad Software, USA). P values of < 0.05 were considered significant. Error bars are expressed as ± SD.
Results
Antiviral peptides design and synthesis
BioMoDroid algorithm was used to design the inhibitory peptides based on the calculation of the the sum of hydrophobic and charge compatibility index for the receptor/interface against the ligand for all possible sequences of the interface. The antiviral peptide sequences were then selected based on the final score, which represented the in silico bonding strength of the receptor-ligand complex. Four antiviral peptide sequences of 10 amino acids that gave the best score for domain III of DENV-2 E protein were selected to test the inhibition of DENV entry and multiplication experimentally. Table 1 shows the amino acid sequences and final BioMoDroid score of the designed peptides.
Evaluation of peptide cytotoxicity
Toxicity was measured to determine the maximum non-toxic dose of the inhibitory peptides. Besides the undesired effect, toxicity could induce cellular alterations that decrease the formation of plaques leading to false interpretation of antiviral activity. Toxic effects ranged from no evidence to minimal toxicity for different peptides when compared to untreated control cells as shown in Figure 1. Results showed that peptide DET3 has the highest toxic effect compared to other peptides and there were no significant differences in terms of time course of activity (Two-way ANOVA with Bonferroni post-test, P > 0.05).
The sequences and properties of the designed and synthesized peptides.
| No | Peptide | Sequence | Length (aa) | BioModDroid Score |
|---|---|---|---|---|
| 1 | DET1 | GWVKPAKLDG | 10 | 0.726262 |
| 2 | DET2 | PWLKPGDLDL | 10 | 0.740630 |
| 3 | DET3 | IGVRPGKLDL | 10 | 0.803902 |
| 4 | DET4 | AGVKDGKLDF | 10 | 0.751608 |
Toxicity of inhibitory peptides in vitro. MTS assay for cell viability was performed with increasing concentration of different peptides at different time courses 24, 48, and 72 hours. Results ranged from no evidence of toxicity to less than 10% for different peptides when compared to untreated control. (A) DET1, (B) DET2, (C) DET3, and (D) DET4. Results are expressed as mean from a representative experiment performed in triplicate.

Evaluation of peptide antiviral activity
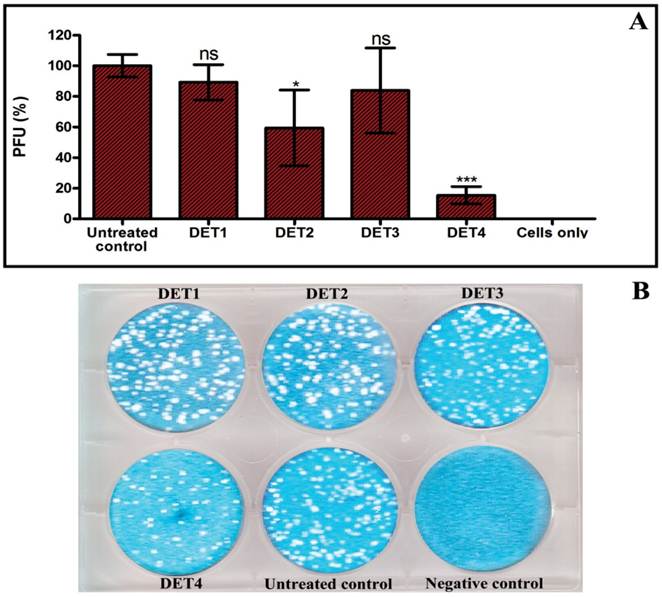
The effectiveness of the designed peptides was verified by testing the antiviral activity of the peptides against DENV-2 using plaque formation assay, RT-qPCR, and Western blot analysis. The antiviral activity analyses were done using the maximal non-toxic dose based on cytotoxicity results of each peptide. In the plaque formation assay, results identified two peptides that were able to inhibit the infection. DET2 and DET4 peptides showed 40.6% ± 24.8 and 84.6% ± 5.6 reduction of plaque formation respectively (One-way ANOVA with Dunnett's post-test, P < 0.0001). Results showed no significant reduction of plaque formation in case of DET1 and DET3 (One-way ANOVA with Dunnett's post-test, P > 0.05) as shown in Figure 2.
These results were further confirmed by quantification of intracellular viral RNA load using RT-qPCR analysis. Results showed that only DET2 and DET4 significantly reduced the DENV entry and, therefore, reduced viral RNA load. The level of reduction was 0.29 fold ± 0.16 (28.6% ± 16.3), and 0.81 fold ± 0.07 (81.0% ± 7.0) for DET2 and DET4 respectively as shown in Figure 3 (One-way ANOVA with Dunnett's post-test, P < 0.0001). DET1 and DET3 did not show significant inhibition of DENV (One-way ANOVA with Dunnett's post-test, P > 0.05).
Evaluation the antiviral activity of the designed peptides by plaque formation assay. The antiviral activity was tested by incubation LLC-MK2 target cells with a mixture of approximately 200 PFU of DENV-2 and 500 µM of each peptide. (A) Histogram shows that DET2 and DET4 are able to inhibit infection by DENV after normalized to untreated control. Results are expressed as mean ± SD from a representative experiment performed in triplicate. Asterisk denotes statistically significant differences from the untreated control (One-way ANOVA with Dunnett's post-test, P < 0.0001). (B) Plaque formation assay shows the reduction of plaque generation.
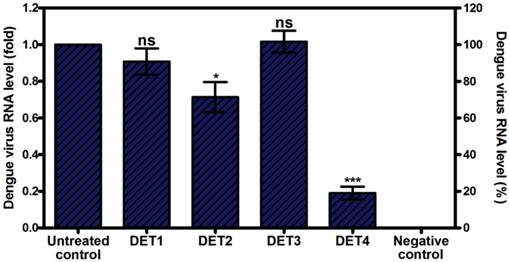
Evaluation the antiviral activity of the designed peptides by RT-qPCR. Viral RNA levels were quantified by RT-qPCR after pre-incubation with antiviral peptides. Data is expressed as relative fold expression compared to untreated control, which was defined as 1.0 fold, and then normalized to β-actin reference gene. Results showed that DET2 and DET4 significantly reduced DENV viral RNA load. The reduction level was 0.29 fold ± 0.16 (28.6% ± 16.3), and 0.81 fold ± 0.07 (81.0% ± 7.0) for DET2 and DET4 respectively. Results are expressed as mean ± SD from a representative experiment performed in quadruple experiments. Asterisk denotes statistically significant differences from the untreated control (One-way ANOVA with Dunnett's post-test, P < 0.0001).
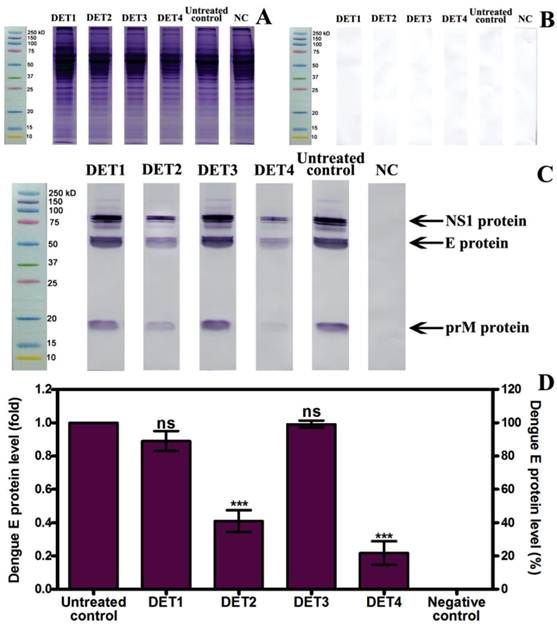
Western blot results showed a significant reduction of the E-protein quantity in cells treated with DET2 and DET4. The percentage of the reduction was 0.59 fold ± 0.11 (59.1% ± 11.2), and 0.78 fold ± 0.12 (78.3% ± 12.2) for DET2 and DET4 respectively as shown in Figure 4 (One-way ANOVA with Dunnett's post-test, P < 0.0001). DET1 and DET3 did not show significant inhibition of DENV (One-way ANOVA with Dunnett's post-test, P > 0.05). It is clear that the immunoblotting was specific as the membrane incubated with dengue immune human serum developed bands (Figure 4C) whereas the membrane incubated with normal human serum did not show any bands (Figure 4B). Furthermore, the bands aligned with the marker at their known molecular weight, namely E-protein of approximately 53 of kDa, NS1 in its dimer form of approximately 90 kDa, and prM of approximately 21 kDa as shown in Figure 4C. The ability of the designed peptides to inhibit DENV was quantified based on the analysis of the E protein densitometry readings because this is a structural protein.
Determination of the peptide inhibitory dose
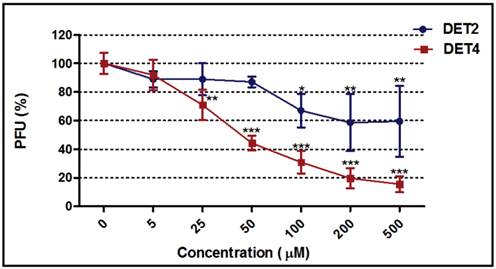
Dose response curves were generated for the most potent peptides (DET2 and DET4) against DENV-2 as shown in Figure 5. The inhibitory activity increased with increasing concentration of both peptides. The DET2 peptide showed a maximum inhibition activity against DENV-2 of 41.5% ± 20.0 at 200 µM with IC50 above 500 µM (One-way ANOVA with Dunnett's post-test, P = 0.0065), while DET4 peptide showed a maximum inhibition activity against DENV-2 of 84.6% ± 5.6 at 500 µM with IC50 of 35 μM (One-way ANOVA with Dunnett's post-test, P < 0.0001).
Evaluation of the post-infection antiviral activity of DET4 peptide
The ability of the DET4 peptide to inhibit DENV-2 after the virus entry into the target cells was determined by quantification of intracellular viral RNA load using RT-qPCR analysis. Peptides were added directly to the cells 24 hours post-infection. Peptide DET4 showed no significant inhibitory effect against DENV-2 when added directly to the target cells 24 hours post-infection as shown in Figure 6 (One-way ANOVA with Dunnett's post-test, P = 0.7697).
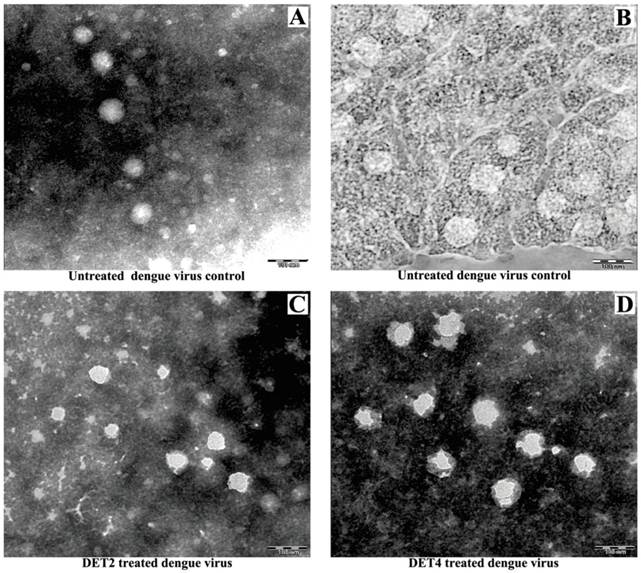
Investigation of the viral surface changes caused by peptide attachment using transmission electron microscope
TEM was used to visualize the effect of treatment DENV-2 with DET2 and DET4 on the viral particles surface. The experiment also includes untreated dengue virions as a control. Results showed that the untreated viral control exhibited the normal-smooth outer surface which is typical for all mature flaviviruses. The surfaces of the virus particles treated with peptides became irregular and had rough edges after treatment with peptides as shown in Figure 7. This may suggest a possible rearrangement of the viral E protein.
Evaluation the antiviral activity of the designed peptides by Western blot. (A) Protein separation by SDS-PAGE. (B) Membrane incubated with normal human serum shows no bands. (C) Membrane incubated with dengue infected human serum develops bands for NS1, E, and prM proteins. (D) Relative quantification of E protein compared to untreated control, which was defined as 1.0 fold, and then normalized to β-actin reference protein. DET2 and DET4 significantly reduced the E protein (0.59 fold ± 0.11, and 0.78 fold ± 0.12 respectively). Results are expressed as mean ± SD from triplicate experiments. Asterisk denotes statistically significant differences from the untreated control (One-way ANOVA with Dunnett's post-test, P < 0.0001).
Dose-response curves for DET2 and DET4. The figure shows the effect of increasing concentrations of the DET2 and DET4 peptides against DENV-2. The DET2 peptide showed a maximum inhibitory activity against DENV-2 of 41.5% ± 20.0 at 200 µM (One-way ANOVA with Dunnett's post-test, P = 0.0065). The DET4 peptide showed a maximum inhibitory activity against DENV-2 of 84.6% ± 5.6 at 500 µM (One-way ANOVA with Dunnett's post-test, P < 0.0001). Results are expressed as mean ± SD from a representative experiment performed in triplicate. Asterisk denotes statistically significant differences from the untreated control.
Evaluation the post-infection antiviral activity of DET4 peptide. The ability of the peptide DET4 to inhibit DENV-2 replication after the virus entry into the target cells was determined by quantification of intracellular viral RNA load using RT-qPCR analysis. Peptides were added to the cells 24 hours post-infection. Then the viral RNA load in peptide treated cells was compared to the untreated control and was normalized to the reference gene (β-actin). Data is expressed as relative percent to untreated control, which was defined as 100%. Peptide DET4 showed no significant inhibitory effect against DENV-2 when added to the infected cells post-infection indicating that the peptide is active through the attachment and entry stage but not viral multiplication. Results are expressed as mean ± SD from a representative experiment performed in triplicate (One-way ANOVA with Dunnett's post-test, P = 0.7697, ns, denotes statistically non-significant differences from the untreated control).

Study the viral surface changes caused by peptide attachment using TEM. DENV was prepared with or without incubation with antiviral peptides before staining with PTA negative stain to visualize by TEM. (A and B) Untreated DENV as a control. (C) DENV treated with DET2 (D) DENV treated with DET4. Scale bars indicate 100 nm.
Discussion
The search for dengue antivirals is a creative endeavor that is gaining momentum due to both increased interest in dengue and substantial progress in the structural biology applications. Since HCV and DENV are members of the same family, Flaviviridae, it is necessary to take advantage of what has been achieved in the drug discovery for HCV and encourage similar strategies to be adopted for DENV such as using RNAi applications [20, 21] and antiviral peptides [22].
DENV entry is a critical step for establishing the infection and a significant inhibition of DENV entry has been achieved by targeting viral entry either using RNAi [23-26] or antiviral peptides [11, 27, 28]. These studies showed that design and synthesis of agents that prevent DENV binding and entry to the cellular receptor sites could prove to be potential antiviral agents for preventing the disease. This study also successfully inhibits DENV entry by targeting the viral E protein with inhibitory peptides. This finding together with previous studies could pave the way to the successful development of therapeutic anti-dengue agents.
Identifying novel peptides with anti-dengue activity could lead to improve the prevention or treatment strategies. The availability of accurate structural data of the DENV E protein and receptor binding sites together with a predictive algorithm could be useful in identifying active inhibitory peptides. There are two possible conditions to design binding peptides, firstly, a peptide is known and the best binding site that provides the optimum binding conditions is explored and secondly, the interface binding surface is given, and a peptide must be designed de novo [29]. In this study, the highly variable region in the lateral loop on domain III that has been implicated in receptor binding of DENV-2 and other flaviviruses such as Tick-Borne Encephalitis Virus [15, 16, 30], and hence was selected as a target for peptide design. Selection of receptor binding site as a target for the antiviral peptides could produce conformational changes or as a physical barrier, to prevent the biological binding of DENV to the cellular surface receptor and, therefore, inhibit viral entry into host cells. There are several successful attempts for computational peptide design using bioinformatics tools and knowledge-based search strategies [31-33]. These tools utilized diverse sets of statistical descriptors, different training databases, different hydrophobicity and charge compatibility indexes, and motif regularities based on known epitopes or protein libraries. In the current study, the peptides were designed based on the iterative and statistical calculations on the amino acid level using hydrophobicity and charge compatibility index between the receptor/interface against the ligand. Although the results were obtained after a series of processing which took almost 15 weeks (with discontinuation), it would not be possible to undertake such hefty manual task of testing each possible peptide sequence in the wet lab not to mention the huge sequencing cost involved. In this case, the peptides were selected based on the final BioMoDroid score, which represented the overall best peptides predicted by the software. This study used a fixed length of 10 amino acids in the design. Future work may attempt in varying the length of the designed peptide or to use and compare other sources of indices and to see the effect in vitro. More effort are required in order to further refine the software techniques as utilized here.
Several previous studies have developed antiviral peptides based on the sequences of the viral E protein. One of these studies designed several inhibitor peptides to inhibit infectivity of DENV by targeting the viral E protein based on Wimley-White interfacial hydrophobicity scale logarithm and its structure [11]. A more recent study reported the use of structural data from the pre-fusion DENV-2 E protein as a model for a computational method to design entry inhibitor peptides [28]. This approach uses the residue-specific all-atom probability discriminatory function (RAPDF) score to identify in situ amino acid sequences that are possible to obtain high binding and structural stability [34, 35].
The antiviral activity that is demonstrated by the two peptides confirms the function of the BioModroid algorithm in designing binding protein ligand. In a previous study, several peptides were designed using known viral structural data with physicochemical algorithm, the Wimley-White interfacial hydrophobicity scale, to inhibit DENV infectivity by targeting viral E protein. One peptide that corresponds to the stem domain of DENV inhibited viral infectivity by reducing 99% of plaque formation in the plaque assay with IC50 in the 10 μM range. This study suggested that this peptide interferes with the intramolecular interactions between the stem and other viral fusion proteins [11]. In another study, a murine brain cDNA phage display library was researched using the E protein of WNV. Numerous adherent peptides that showed significant binding to WNV E protein were selected for in vitro antiviral activity evaluation. Two peptides significantly inhibited the WNV infection and one of them showed an inhibition activity against DENV with IC50 range of 67 μM [27]. In a more recent study, multiple inhibitor peptides were designed using computational predictive strategies together with high-resolution structural data of DENV E protein. Again two peptides showed inhibition of viral entry by interfering with virus binding to the host cells surface [28]. Similarly, other peptides derived from the stem of DENV E protein showed blocking of viral fusion and inhibition of infectivity. The activity of these peptides depend upon their affinity for the conformational intermediate because they bind to the viruses before attachment and are carried with virions into endosomes where the acidification initiates the fusion [12]. The results of the current study are consistent with the finding of the previous studies that showed inhibition of DENV entry using micromolar range of peptides concentration although, it targets a different region of the DENV E protein. The present study was unable to demonstrate the effectiveness of the designed peptides with other flaviviruses due to the deficiency of necessary facilities. Hence, the peptides designed in this study can function as lead compounds for developing antiviral drugs against the DENV and potentially other flaviviruses.
This study proposed that these active inhibitor peptides inhibit DENV entry by blocking biological attachment and causing structural rearrangements of the viral E protein, which are important for establishing the viral infection. The non-inhibitory effect seen by adding the peptides cells post-infection indicates that the DET4 peptide is active through the attachment or viral entry stage in the virus life cycle and not once virus has entered the cell.
Mature DENV is similar to other flaviviruses in that it exhibits a normal and smooth outer surface [36]. Inhibitory peptides caused changes in the surface of viruses from smooth to rough compared to the untreated controls and non-inhibitory peptides. The TEM images show that these antiviral peptides could cause structural abnormalities in infectious virions. These images do not provide any details about the kinetics of these modifications. This result indicates the possibility of the presence of variation in the viral surface E protein arrangement, which interferes with virus binding and entry into host cells. However, it doesn't exclude that such changes could be deleterious to the virus and may be a virucidal effect. These data are consistent with previous studies that showed the mutations of the domain III of flaviviruses can reduce their virulence [37-40]. Furthermore, a previous study carried out by Costin et al showed that the peptides with antiviral activity also showed changes to the viral surfaces as revealed by cryo-electron microscope analysis [28].
Conclusions
The incidence of fatal dengue hemorrhagic fever cases has increased sharply especially in Asia over the last two decades, making it a leading cause of morbidity by inducing coagulopathy and vasculopathy [41]. One of the principal virological characteristics of severe dengue hemorrhagic fever patients is the elevated viral load even during the defervescence stage [42]. Taken together, the outcomes of this study support the hypothesis that these active peptides interact directly with DENV-2 E proteins and interfere with viral entry. Domain III of the envelope protein in dengue viruses is a target of both serotype-specific and dengue cross-reactive neutralizing antibodies and there are several conserved and variable regions between the four serotypes. DET2 and DET4 were designed to target the short sequence (residues 380-IGVEPGQLKL-389) in the lateral loop on domain III of DENV-2 envelope protein which has been implicated in receptor-binding. This region is variable among dengue serotypes but conserved within DENV2 strains. This variability effect on peptide activity needs further in vitro evaluation against the other dengue virus serotypes. These inhibitory peptides against DENV-2 could act as the basis for designing and developing novel therapeutics for dengue infection by all the four serotypes. Disparate chronic infections, immune responses against DENV and other flaviviruses have the ability to clear the viruses from the individuals that persist to the primary infection. Reducing the viral load during the initial stages of infection can increase the window of time to raise the immune response and therefore allow more individuals to eliminate, control, and survive infections by these agents.
Abbreviations
DENV: Dengue virus; E protein: envelope protein; TEM: Transmission electron microscopy; HSV: Herpes simplex virus; LLC-MK2: Rhesus Monkey Kidney Epithelial Cells; DMEM: Dulbecco's Modified Eagle Medium; FBS: Fetal bovine serum; HPC: High performance computing; PFU: Plaque forming units; PBS: Phosphate buffered saline; RT-qPCR: Reverse transcription quantitative real-time PCR; HCV: Hepatitis C Virus; IC50: Half maximal inhibitory concentration; WNV: West Nile Virus
Acknowledgements
Thanks are addressed to Electron Microscopy Unit, Faculty of Medicine, University of Malaya for using TEM facility.
Funding
This project was supported by High Impact Research Grant from University Malaya J-00000-73560, H-200001-00-E000079 and University of Malaya IPPP Grant: PV053/2011B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen Y, Maguire T, Marks RM. Demonstration of binding of dengue virus envelope protein to target cells. J Virol. 1996;70:8765-72
2. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717-25
3. Clyde K, Kyle JL, Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J Virol. 2006;80:11418-31
4. Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313-9
5. Hancock RE, Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends in microbiology. 2000;8:402-10
6. Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clinical Microbiology Reviews. 2006;19:491-511
7. Benincasa M, Skerlavaj B, Gennaro R, Pellegrini A, Zanetti M. In vitro and in vivo antimicrobial activity of two alpha-helical cathelicidin peptides and of their synthetic analogs. Peptides. 2003;24:1723-31
8. Albiol Matanic VC, Castilla V. Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int J Antimicrob Agents. 2004;23:382-9
9. Yasin B, Wang W, Pang M, Cheshenko N, Hong T, Waring AJ. et al. Theta defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. Journal of virology. 2004;78:5147-56
10. Yasin B, Pang M, Turner JS, Cho Y, Dinh NN, Waring AJ. et al. Evaluation of the inactivation of infectious Herpes simplex virus by host-defense peptides. Eur J Clin Microbiol Infect Dis. 2000;19:187-94
11. Hrobowski YM, Garry RF, Michael SF. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol J. 2005;2:49
12. Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 2010;6:e1000851
13. Das S, Garver L, Ramirez JR, Xi Z, Dimopoulos G. Protocol for Dengue Infections in Mosquitoes (A. aegypti) and Infection Phenotype Determination. J Vis Exp. 2007:e220
14. De Madrid AT, Porterfield JS. A simple micro-culture method for the study of group B arboviruses. Bulletin of the World Health Organization. 1969;40:113
15. Hung JJ, Hsieh MT, Young MJ, Kao CL, King CC, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. Journal of virology. 2004;78:378-88
16. Mazumder R, Hu ZZ, Vinayaka CR, Sagripanti JL, Frost SD, Kosakovsky Pond SL. et al. Computational analysis and identification of amino acid sites in dengue E proteins relevant to development of diagnostics and vaccines. Virus Genes. 2007;35:175-86
17. Biro JC. Amino acid size, charge, hydropathy indices and matrices for protein structure analysis. Theoretical biology & medical modelling. 2006;3:15
18. Yang JM, Chen YF, Tu YY, Yen KR, Yang YL. Combinatorial computational approaches to identify tetracycline derivatives as flavivirus inhibitors. PLoS One. 2007;2:e428
19. Kong YY, Thay CH, Tin TC, Devi S. Rapid detection, serotyping and quantitation of dengue viruses by TaqMan real-time one-step RT-PCR. Journal of Virological Methods. 2006;138:123-30
20. Nakagawa S, Umehara T, Matsuda C, Kuge S, Sudoh M, Kohara M. Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochemical and Biophysical Research Communications. 2007;353:882-8
21. Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P. et al. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413-21
22. Paeshuyse J, Kaul A, De Clercq E, Rosenwirth B, Dumont JM, Scalfaro P. et al. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761-70
23. Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H. et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242-5
24. Subramanya S, Kim SS, Abraham S, Yao J, Kumar M, Kumar P. et al. Targeted delivery of small interfering RNA to human dendritic cells to suppress dengue virus infection and associated proinflammatory cytokine production. J Virol. 2010;84:2490-501
25. Alhoot MA, Wang SM, Sekaran SD. Inhibition of Dengue Virus Entry and Multiplication into Monocytes Using RNA Interference. PLoS Negl Trop Dis. 2011;5:e1410
26. Alhoot MA, Wang SM, Sekaran SD. RNA Interference Mediated Inhibition of Dengue Virus Multiplication and Entry in HepG2 Cells. PLoS One. 2012;7:e34060
27. Bai F, Town T, Pradhan D. et al. Antiviral peptides targeting the west nile virus envelope protein. J Virol. 2007;81:2047-55
28. Costin JM, Jenwitheesuk E, Lok SM, Hunsperger E, Conrads KA, Fontaine KA. et al. Structural optimization and de novo design of dengue virus entry inhibitory peptides. PLoS Negl Trop Dis. 2010;4:e721
29. Unal EB, Gursoy A, Erman B. VitAL: Viterbi algorithm for de novo peptide design. PLoS One. 2010;5:e10926
30. Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J. et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728-38
31. Ying C, De Clercq E, Neyts J. Selective Inhibition of Hepatitis B Virus Replication by RNA Interference. Biochemical and Biophysical Research Communications. 2003;309:482-4
32. McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H. et al. Inhibition of Hepatitis B Virus in Mice by RNA Interference. Nature biotechnology. 2003;21:639-44
33. Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small Interfering RNA Inhibits Hepatitis B Virus Replication in Mice. Molecular Therapy. 2003;8:769-76
34. Huang ES, Samudrala R, Park BH. Scoring functions for ab initio protein structure prediction. Methods Mol Biol. 2000;143:223-45
35. Samudrala R, Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J Mol Biol. 1998;275:895-916
36. Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S. et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nature structural & molecular biology. 2008;15:312-7
37. Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, Ramos C. et al. Dengue virus structural differences that correlate with pathogenesis. Journal of virology. 1999;73:4738
38. Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T. et al. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. Journal of virology. 2001;75:5627-37
39. Sánchez IJ, Ruiz BH. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. Journal of general virology. 1996;77:2541
40. Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ. et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866-71
41. Ho LJ, Wang JJ, Shaio MF, Kao CL, Chang DM, Han SW. et al. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J Immunol. 2001;166:1499-506
42. Wang WK, Chao DY, Kao CL, Wu HC, Liu YC, Li CM. et al. High levels of plasma dengue viral load during defervescence in patients with dengue hemorrhagic fever: implications for pathogenesis. Virology. 2003;305:330-8
Author contact
![]() Corresponding author: E-Mail: shamalamycom. Address: Department of Medical Microbiology, Faculty of Medicine, University of Malaya, Lembah Pantai, 50603 Kuala Lumpur, Malaysia. Tel: +603 7967 5759. Fax: +603 7967 6672.
Corresponding author: E-Mail: shamalamycom. Address: Department of Medical Microbiology, Faculty of Medicine, University of Malaya, Lembah Pantai, 50603 Kuala Lumpur, Malaysia. Tel: +603 7967 5759. Fax: +603 7967 6672.

 Global reach, higher impact
Global reach, higher impact