Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(5):498-507. doi:10.7150/ijms.5560 This issue Cite
Research Paper
PML(NLS-) Inhibits Cell Apoptosis and Promotes Proliferation in HL-60 Cells
1. Central Laboratory of Yong-chuan hospital, Chongqing Medical University, Chongqing 402160, China.
2. Key Laboratory of Laboratory Medical Diagnostics, Ministry of Education, Department of Laboratory Medicine, Chong-qing Medical University, Chongqing 400016, China.
Received 2012-11-17; Accepted 2013-2-22; Published 2013-3-5
Abstract
Promyelocytic leukemia (PML) is a cell-growth suppressor, and PML-retinoic acid receptor α (PML-RARα) is known as a fusion gene of acute promyelocytic leukemia (APL). Studies have reported that neutrophil elastase(NE) cleaved bcr-1-derived PML-RARα in early myeloid cells leading to the removal of nuclear localization signal (NLS) from PML. The resultant PML without NLS named PML(NLS-). PML(NLS-) mainly locates and functions in the cytoplasm. PML(NLS-) may act in different ways from PML, but its biological characteristics have not been reported. In this study, the PML (NLS-) was silenced with shRNA [HL-60/pPML(NLS-)-shRNA] and over-expressed by preparation of recombinant adenovirus [HL-60/pAd-PML(NLS-)]. The mRNA and protein expression of PML(NLS-) were detected by RT-PCR and Western blot respectively. Cell proliferation in vitro was assessed by MTT assay. Flow cytometry (FCM) was used to detect apoptotic cells. The transcription of BCL-2, BAX and C-MYC was detected in HL-60/pAd-PML(NLS-) cells. Our results showed that compared to the control group, the expression of PML(NLS-) was significantly reduced in the HL-60/pPML(NLS-)-shRNA cells, and increased significantly in the HL-60/pAd-PML(NLS-) cells. The proliferation was significantly inhibited in the HL-60/pPML(NLS-)-shRNA cells in a time-dependent manner, but markedly promoted in the HL-60/pAd-PML(NLS-) cells treated with 60 μmol/L emodin. FCM revealed the apoptosis increased in HL-60/pPML(NLS-)-shRNA cells, and decreased in the HL-60/pAd-PML(NLS-) cells. The expression of BAX decreased significantly, while that of BCL-2 and C-MYC increased significantly in HL-60/ pAd-PML(NLS-) cells. Down-regulation of PML(NLS-) expression inhibits the proliferation and induces the apoptosis of HL-60 cells. On the contrary, over-expression of PML(NLS-) promotes the proliferation and reduce the emodin-induced apoptosis of HL-60 cells.
Keywords: PML(NLS-), shRNA, over-expression, proliferation, apoptosis.
Introduction
Promyelocytic leukemia (PML), also known as “PML NBs”, “ND10”, “Kr bodies”, “PODs” and “PML bodies”[1], is encoded by PML gene mapped on chromosome 15q22 in humans [2]. The full length of PML gene is about 53147 bp. The PML bodies consist of at least 15 components [3], and are dynamic macromolecular multiprotein complexes that can recruit and release a plethora of proteins [4]. The amount and size of varies throughout the cell cycle. The PML nuclear bodies (NB) are the lowest in amount in the G0 phase then slowly increase during the progression to G1 phase, and are the highest in amount in the S phase [5, 6]. The PML NB components play vital roles in the regulation of multiple cellular functions such as apoptosis, senescence, tumor suppression, transcription, DNA repair, and proteolysis [7]. The PML protein exists in different isoforms, which vary in size from 47 kD to 160 kD, are generated by alternative splicing and have variable C-terminal lengths [8]. However, all the isoforms contain nuclear localization signal (NLS), B-Boxes and α-helical coiled-coil region [9].
PML gene on 15q22 fuses with a retinoic acid receptor alpha (RARα) gene on 17q21 giving rise to a PML-RARα gene fusion product [10]. Some studies have shown that the transgenic and knock-in animals expressing PML-RARα in early myeloid cells [11, 12, 13] developed acute promyelocytic leukemia (APL), but APL was absent when PML-RARα was expressed in late myeloid cells [14]. However, the mechanisms by which PML-RARα predisposes early myeloid cells to eventual leukemic transformation are not yet completely understood. Recently, our results showed neutrophil elastase (NE), an early myeloid-specific serine protease, is important for the development of APL in mice. NE can cleave bcr-1 derived PML-RARα protein in early myeloid cells [15] resulting in removal of NLS from PML. The resultant PML without NLS was named as PML(NLS-). The PML(NLS-) gene is about 1268 bp in length and encodes a protein of 53 kD. To date, the biological functions of PML(NLS-) have not been reported.
In order to investigate the biological characteristics of PML (NLS-) gene, the PML(NLS-) was silenced with shRNA and over-expressed by preparation of adenovirus vector. It has been reported that emodin at 60 μmol/L can effectively inhibit the proliferation of APL cell line (HL-60 cells) and induce their apoptosis[16]. Thus, HL-60 cells were employed and transfected with recombinant adenovirus carrying PML (NLS-) and treated with 60 μmol/L emodin. The effects of PML (NLS-) on emodin-induced proliferation and apoptosis were investigated in HL-60 cells.
Materials and methods
Cell line and culture
Human HL-60 cells were purchased from the Shanghai Institute for Biological Science and maintained in IMDM (Gibco, MD, USA) containing 20% fetal bovine serum (FBS; Gibco, MD, USA) in an environment with 5% CO2 at 37 °C.
Transient transfection
HL-60 cells (2.5×105/ml) in logarithmic growth phase were seeded in 6-well plate. For transfection, 2.5 μg of plasmid pPML(NLS-)-shRNA was diluted in 500 μl Opti-MEM I, which was added to each well. Then, 2.5 μl of PLUS™ Reagent was directly added to dilute DNA. After gentle mixing and incubation for 15 min at room temperature, 13.75 μl of Lipofectamine™ LTX was added into above DNA solution, followed by incubation for 25 min at room temperature. The resultant solution was added to each well. After incubation at 37°C for 6 h, 1 ml of fresh complete medium was added. These transfected cells were named as HL-60/pPML(NLS-)-shRNA. In the negative control group, HL-60/pshRNA-NC cells were processed similarly. At 48 h after transfection, the successfully transfected cells were verified by fluorescent microscopy. The transfection efficiency was expressed as the percentage of GFP-positive cells. There were three groups in this experiment: HL-60 cells group, HL-60/pshRNA-NC group and HL-60/pPML(NLS-)-shRNA group.
Cell infection by adenovirus
HL-60 cells in logarithmic growth phase (5×105/well) were seeded in 6-well plate. These cells were infected with RFP-expressing adenovirus vector pAd-KZ and PML(NLS-)-expressing recombinant adenovirus pAd-PML(NLS-). Meanwhile, an appropriate amount of polybrene was added. After culture for 8-12 h, the medium was refreshed. The fluorescence was then detected 48 h later. The recombinant HL-60/pAd-PML(NLS-) cells were used for following experiments. There were three groups in this experiment: HL-60 cells group, HL-60/pAd-KZ groups, and HL-60/pAd-PML(NLS-) group.
RNA isolation and RT-PCR
Total RNA was extracted from cells in each group using Trizol reagent (Invitrogen, Carlsbad, California), and RT-PCR was performed using a PreTaq kit (TaKaRa, Dalian, China) and a Quantity One Software (BIO-RAD, USA). In brief, 1 μg of RNA was reversibly transcribed to cDNA in 10-μl solution with the ThermoScript™RT-PCR system (Invitrogen Life Technologies, Inc, Carlsbad, CA, USA).
The reaction mixture contained 12 μl of PreTaq Enzyme, 0.5 μl (10μM) of each primer and 100 ng of cDNA in a 25-μl solution. PCR conditions were as follows: pre-denaturation at 95°C for 5 min, 35 cycles of denaturation at 95 °C for 30 s, annealing at 64 °C for 30 s, and extension at 72 °C for 75 s, and a final extension at 72 °C for 5 min. The amplification of β-actin gene was the same to that of PML(NLS-). Both PCR products were electrophoresed on 1% agarose gel and stained with gold view. The mRNA expression of PML(NLS-) was normalized to that of β-actin.
Western blot assay
Cells in each group were washed with ice-cold phosphate-buffered saline and lysed in RIPA solution containing protease inhibitor cocktail. Protein concentration was determined with BCA method. A total of 100 μg of protein was added in 12% sodium dodecyl sulfate-polyacrylamide gel, and then transferred to polyvinylidene difluoride membrane. The member was blocked in 5% non-fat milk for 4 h, then incubated with primary antibody overnight at 4 °C (rabbit polyclonal antibody against PML; 1:2000; Abcam, Hong Kong, China) and then with secondary antibody (goat anti-rabbit antibody, 1:1000, Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China) for 1 h at 37 °C. After washing with Tris-Buffered Saline Tween-20 (TBST), the autoradiograms were scanned and subjected to densitometry. β-actin (mouse monoclonal antibody against β-actin, 1:500; Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China) was used as an internal control.
Cell viability assay
To assess cell viability, cells in each group were seeded in 96-well plates at a density of 1.0×104 cells/well. Cell proliferation was quantified in 4 days by MTT assay. In brief, 20 μl of MTT (5 mg/ml; Sigma, MO, USA) was added to each well followed by incubation for 4 h at 37°C. The medium was then replaced with 150 μl of dimethylsulphoxide (DMSO, Sigma, MO, USA). The cell viability was assessed by detection of absorbance at 492 nm using a spectrophotometer. Cells growth curves were plotted. The experiment was repeated at least three times.
The main difference is in need of adding emodin to evaluate the cell proliferation ability of adenovirus groups. In the emodin treated group, HL-60 cells were seeded in 96-well plates (1.0×104/well). Recombinant adenovirus pAd-PML(NLS-) mixture and pAd-KZ mixture containing polybrene were independently added into each well after culture for 8-12 h. At the same time, 60 μmol/L emodin was added into each well. These cells were named as HL-60/pAd-PML(NLS-) group and HL-60/pAd-KZ group, respectively. In the blank control group, cells were only treated with 60 μmol/L emodin. After culture for 1-4 days, the cell viability was measured by MTT assay as above mentioned.
Flow cytometry
Cells in transfection group were harvested by centrifugation at 2000 rpm for 5 min. After washing twice with pre-cold PBS (pH 7.4), cells were re-suspended in Binding Buffer, and then stained by Annexin V-PE and 7-AAD for 5-15 min at room temperature. At last, these cells were subjected to flow cytometer (Becton Dickinson, CA, USA) for detection of apoptotic cells. Each experiment was repeated at least three times.
To preferably evaluate the apoptosis level of infection group, besides the treatment of polybrene and a variety of adenoviruses, 60 μmol/L emodin was added into each well in each group. After incubation for 48h, cells of each group were processed as above mentioned. The difference was that cells were stained with single dye Annexin V-FITC.
Detection of expression of apoptosis-related genes by RT-PCR and Western blot assay
The extraction of total RNA and reverse transcription into cDNA were the same to above mentioned. The primers are shown in Table 1. PCR condition was as follow: pre-denaturation at 95°C for 5 min; 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s and extension at 72°C for 50 s; a final extension at 72°C for 5 min. The products were electrophoresed on 1.5% agarose gel and analyzed with Quantity One Software.
Statistical analysis
Data was expressed as means ± standard deviation (SD). Statistical analysis was performed with SPSS version 16.0. Independent sample t test was employed for comparing the means between two groups. A value of P<0.05 was considered statistically significant.
The primers.

Results
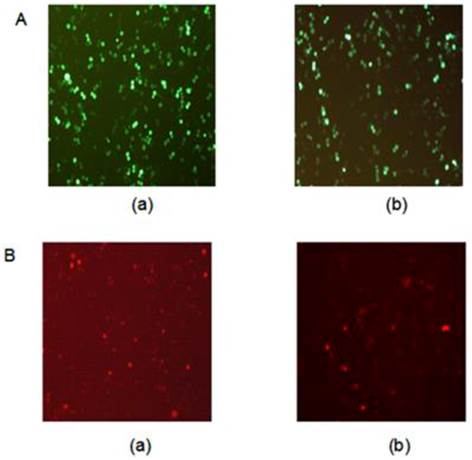
Fluorescent protein expression in HL-60 cells
In this study, pPML(NLS-)-shRNA was introduced in HL-60 cells to silence PML(NLS-) expression. The GFP-positive cells were >70% at 48 h after transfection (Figure 1A).
Meanwhile, recombinant adenovirus pAd-PML(NLS-) was used to infect HL-60 cells to induce PML(NLS-) over-expression. The percentage of GFP-positive cells was about 40% at 48 h, and about 70% at 72 h after infection (Figure 1B).
A, GFP protein expression in transfected cells was detected by fluorescence microscopy (×100). (a) HL-60/pshRNA-NC cells; (b) HL-60/pPML(NLS-)-shRNA cells. B, GFP protein expression in infected cells was detected by fluorescence microscopy (×100). (a) HL-60/pAd-KZ cells; (b) HL-60/pAd-PML (NLS-) cells.
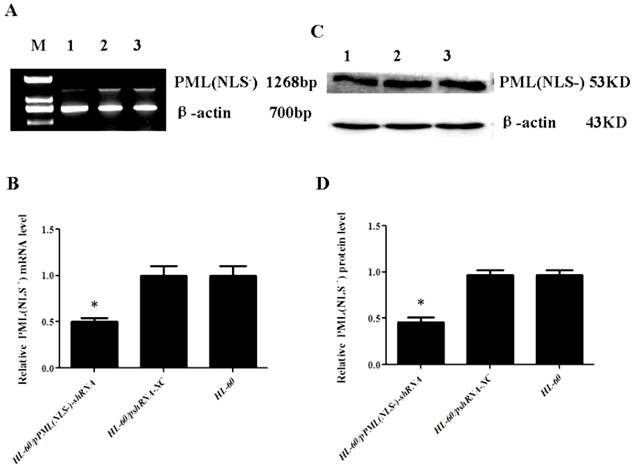
Down-regulation of PML(NLS-) in HL-60/pPML(NLS-)-shRNA cells
RT-PCR and Western blot assay showed that the mRNA and protein expressions of PML(NLS-) were significantly lowered in the HL-60/pPML(NLS-) -shRNA cells when compared with control group (P<0.05, Figure 2). No significant difference in the mRNA and protein expressions of PML(NLS-) was found between HL-60 cells and HL-60/pshRNA-NC cells (P>0.05, Figure 2).
A and B, mRNA expression of PML(NLS-) was detected by RT-PCR. The mRNA expression of PML(NLS-) in the HL-60/pPML(NLS-)-shRNA cells was significantly decreased when compared with control group. Data was expressed as means ±SD. *P<0.05. C and D, Protein expression of PML(NLS-) was assessed by Western blot assay. The protein expression of PML(NLS-) in the HL-60/pPML(NLS-)-shRNA cells was significantly decreased when compared with the controls. Data was expressed as means±SD. * P<0.05 (lane 1: HL-60/pPML(NLS-)-shRNA cells; lane 2: HL-60/pshRNA-NC cells; lane 3: HL-60 cells ).
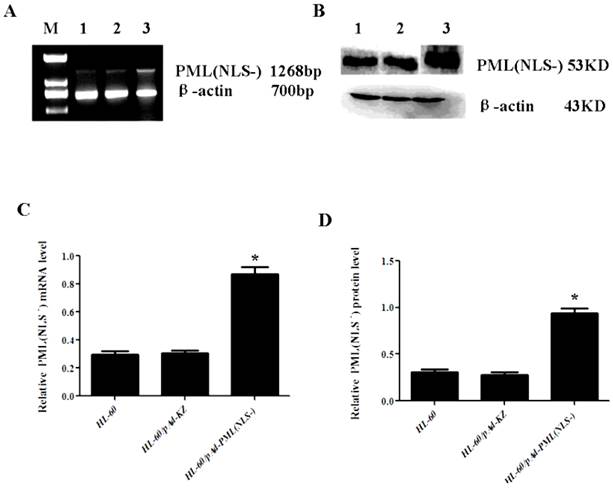
Up-regulation of PML(NLS-) in HL-60/pAd-PML(NLS-) cells
The mRNA and protein expressions of PML(NLS-) were also detected in cells with PML(NLS-) over-expression by RT-PCR and western blot assay. Results revealed that the PML(NLS-) expressions in the HL-60/pAd-PML(NLS-) cells were significantly higher than those in the control groups respectively (P<0.05, Figure 3).
A and C, mRNA expression of PML(NLS-) was detected by RT-PCR. The relative mRNA expression of PML(NLS-) in the HL-60/pAd-PML(NLS-) cells was significantly increased when compared with the control group. Data was expressed as means ± SD. *P<0.05. B and D, The protein expression of PML(NLS-) protein was assessed by Western blot assay. The protein expression of PML(NLS-) in the HL-60/pAd-PML(NLS-) cells was significantly decreased when compared with controls. Data was expressed as means ± SD. *P<0.05 (lane 1: HL-60/pAd-PML(NLS-) cells; lane 2: HL-60/pAd-KZ cells; lane 3: HL-60 cells).
Fluorescent protein expression in HL-60 cells of each group.
Expression of PML(NLS-) in transfected cells.
Expression of PML(NLS-) in infected cells.
Effect of PML(NLS-) on proliferation of HL-60 cells
MTT assay showed that, when compared with the HL-60/pshRNA-NC cells, the proliferation of HL-60/pPML(NLS-)-shRNA cells was significantly inhibited in a time-dependent manner and the highest inhibition rate was 47± 2% at 48 h (P<0.05, Figure 4A). In addition, up-regulation of PML(NLS-) could promote the proliferation of emodin-treated HL-60 cells (Figure 4B).
A, MTT assay was used to detect the proliferation of transfected cells. On days 1, 2, 3 and 4, the absorbance was measured at 492 nm. The proliferation of pPML(NLS-)-shRNA cells was significantly inhibited in a time-dependent manner. Data was expressed as means ± SD. *P<0.05 (lane 1: HL-60/pPML(NLS-)-shRNA cells; lane 2: HL-60/pshRNA-NC cells; lane 3: HL-60 cells). B, MTT assay was used to detect the proliferation of infected cells. Results showed the proliferation of HL-60/pAd-PML(NLS-) cells reduced significantly when compared with controls. Data was expressed as mean ± SD. * P<0.05 (lane 1: HL-60/pAd-PML(NLS-) cells; lane 2: HL-60/pAd-KZ cells; lane 3: HL-60 cells).
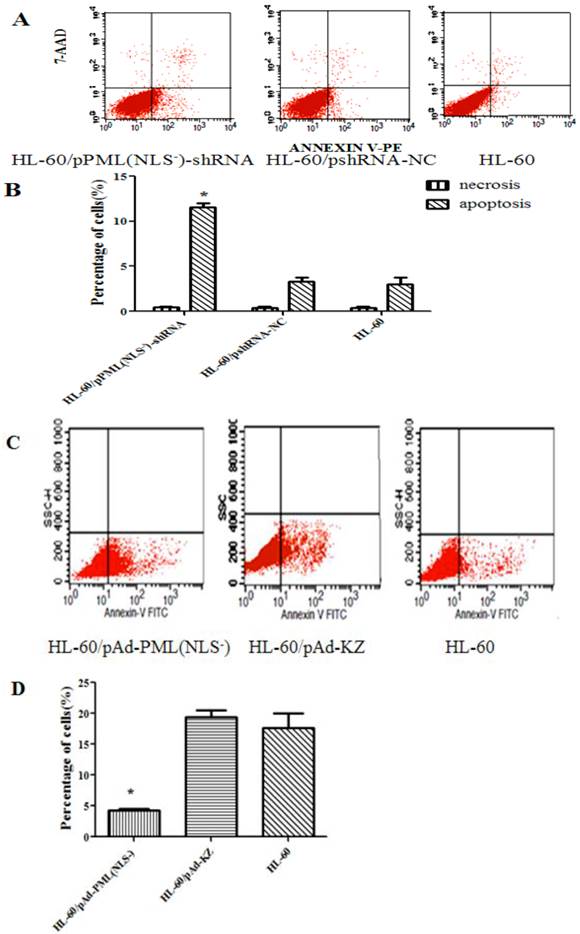
Cell apoptosis
As shown in Figure 5, the apoptosis rate of HL-60/pPML(NLS-)-shRNA group (11.14±0.68%) was significantly higher than that of the control groups (3.77±0.87% and 2.26±0.78%) after transfection for 48 h (P < 0.05). The apoptosis rate of HL-60/pAd-PML(NLS-) group (4.32±0.68%) was markedly lower than that of the control groups (15.00±0.87% and 19.98±0.78%) after treatment for 48 h (P < 0.05). These results indicated that PML(NLS-) could promote the proliferation of emodin-treated HL-60 cells through regulating cell apoptosis.
A and B, The apoptosis rate of transfected HL-60 cells was detected by FCM. The apoptotic HL-60/pPML(NLS-)-shRNA cells increased significantly (*P< 0.05).
C and D, The apoptosis rate of infected HL-60 cells was detected by FCM. The apoptotic HL-60/pAd-PML(NLS-) cells reduced dramatically (*P< 0.05).
Expressions of apoptosis-related gene in HL-60/ pAd-PML(NLS-) cells
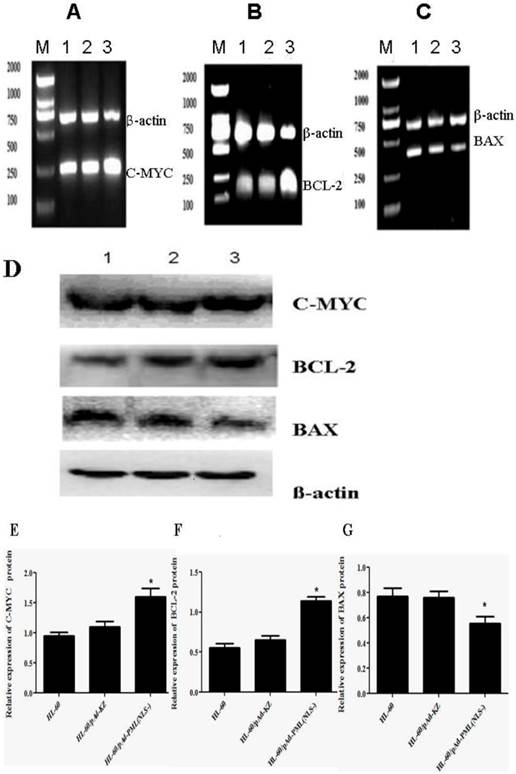
To further investigate the effect of PML(NLS-) on the apoptosis of HL-60 cells, the expressions of apoptosis-related genes (C-MYC, BAX and BCL-2) were detected by RT-PCR and Western blot assay. Results showed that, when compared with controls, the mRNA and protein expressions of C-MYC and BCL-2 in HL-60/pAd-PML(NLS-) cells increased markedly (P<0.05, Figure 6A, B, E, F), and the mRNA and protein expressions of BAX decreased significantly (P<0.05, Figure 6C, G). However, there were no significant differences in the expressions of C-MYC, BCL-2 and BAX between HL-60 cells and HL-60/pAd-KZ cells (P>0.05, Figure 4A, B). These findings suggested that PML(NLS-) could promote the expression of C-MYC and BCL-2 and inhibit that of BAX.
A, B and C, Detection of mRNA expression of C-MYC, BCL-2 and BAX by RT-PCR. When compared with control groups, the expressions of C-MYC and BCL-2 increased significantly (P<0.05), but that of BAX decreased markedly (p<0.05). D, E, F and G, Detection of protein expression of these genes by Western blot assay. The changes in the protein expression of these genes were consistent with those in mRNA expression (lane 1: HL-60 cells; lane 2: HL-60/pAd-KZ cells; lane 3: HL-60/pAd-PML(NLS-) cells ).
Proliferation of HL-60 cells in different groups.

Apoptosis rate in different cells.
Expression of apoptosis-related genes.
Discussion
The fusion gene PML-RARα is the marker of APL and plays an important role in the occurrence and development of APL. Its importance has been confirmed in transgenic mice. However, there is evidence showing that PML-RARα is not always as a whole to play a role in the development of APL [17, 18]. Studies have shown that PML-RARα protein can be cleaved by NE in early myeloid cells and the dominant cleavage sites have been identified within the carboxy-terminal domain of PML in the fusion protein. As a result, this cleavage leads to the separation of NLS from the PML resulting in formation of PML without of NLS, a new mutant of PML, which was named as PML(NLS-). Whether the PML(NLS-) has the same functions to the PML of wild-type is still unclear. In addition, the effect of PML(NLS-) on leukemia cells is poorly understood.
Human wild-type PML acts as a cell-growth suppressor [19, 20, 21, 22] and has different characteristics from PML(NLS-). At present, our findings demonstrated that PML(NLS-) could promote the proliferation of leukemic cells and inhibit their apoptosis.
The PML(NLS-) was silenced with shRNA and over-expressed after recombinant adenovirus infection. RNA interference is a gene silencing process mediated by double-stranded RNA, and an effective method for gene silencing. In order to select specific and optimal target sequence, three pairs of sequences targeting PML(NLS-) were designed, and the interference efficiency of each pair was detected by real-time PCR and western blot assay (data not shown). The most effective one was selected for further studies.
Leukemia cells belonged to suspension cells, therefore we used adenovirus vector carrying expressing PML(NLS-) gene to infect leukemia cell lines (HL-60 cells) for achieving the effect of high infection rates, and target gene was expression steadily expressed in these cells. The recombinant adenovirus pAd-PML(NLS-) was constructed previously (saved in our laboratory), which was successful to carry the PML(NLS-) gene. As shown in Figure 2 and Figure 3, the PML(NLS-) was successfully silenced and over-expressed.
Then, the in vitro proliferation and apoptosis of leukemic cells were investigated in cells with down-regulation or up-regulation of PML(NLS-) expression. MTT assay showed that the proliferation of HL-60/pPML(NLS-)-shRNA was significantly inhibited in a time-dependent manner, but that of HL-60/pAd-PML(NLS-) was markedly promoted when compared with controls. These findings indicated that PML(NLS-) could promote the proliferation of HL-60 cells, which was different from the wild-type PML. In addition, when compared with controls, the apoptosis rate of HL-60/pPML(NLS-)-shRNA group increased significantly, but the necrosis rate among three groups was comparable. However, the apoptosis rate of HL-60/pAd-PML(NLS-) group decreased significantly. These findings showed that over-expression of PML (NLS-) could promote the proliferation of APL cells treated with 60 μmol/L emodin and inhibit their apoptosis, which was different from the biological effect of PML. The wild-type PML inhibited the cell growth and promoted apoptosis. We speculated that this difference could be associated with different orientation of both proteins. Wild-type PML with NLS distributes in the PML nuclear body in the nucleus where tumor suppressor p53 [18], Daxx, p300 [23] and others localize and can interact with it. However, because of lack of NLS, mutant PML localizes in the cytoplasm. We hypothesize that the ectopic PML(NLS-) may fail to interact with these proteins, exerting different biological effects from wild-type PML.
In order to determine the effect of PML(NLS-) on the apoptosis of HL-60 cells, the mRNA and protein expression of apoptosis-related genes were detected. Apoptosis is a type of programmed cell death triggered by some physiological or pathological factors, mainly in the death receptor way and mitochondrial way. BCL-2 family is one of members to regulate apoptosis [24] and often used as a target in the tumor treatment [25]. Among the apoptosis-related genes, BAX and BCL-2 are a pair of genes having opposite effects. The ratio of BAX to BCL-2 is related to the apoptosis. The RT-PCR and Western blot assay showed that over-expression of PML(NLS-) could reduce the BAX expression and increase the BCL-2 expression. As a result, the BAX/BCL-2 ratio decreased. As a bidirectional apoptosis-regulatory gene, C-MYC can promote the proliferation and inhibit the apoptosis when it is over-expressed in pathological conditions. Results also showed that the over-expression of PML(NLS-) could inhibit apoptosis by increasing the expressions of BCL-2 and C-MYC, and decreasing the expression of BAX. However, further studies are required to elucidate the molecular mechanism underlying the effects of PML(NLS-) on the apoptosis of leukemic cells.
Conclusion
Our findings suggest that down-regulation PML(NLS-) expression can inhibit the proliferation of HL-60 cells and induce their apoptosis. PML(NLS-) is a putative tumor promoter factor in human leukemia cells. PML(NLS-) over-expression can promote the growth and induce the apoptosis. PML(NLS-) may become a potential target in the therapy of leukemia. Further investigations may be helpful to develop novel therapeutic approaches for leukemia.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81171658) and Natural Science Foundation Project of CQ CSTC (No. 2011BA5037).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Doucas V, Evans RM. The PML nuclear compartment and cancer. Biochim Biophys Acta. 1996;1288:25-9
2. Lane AA, Ley TJ. Neutrophil Elastase Is Important for PML-Retinoic Acid Receptor Activities in Early Myeloid Cells. Molecular and Cellular Biology. 2005;125:23-33
3. Maria H, Catherine T, Kathy H. et al. Structure, Organization, and Dynamics of Promyelocytic Leukemia Protein Nuclear Bodies. Am. J. Hum. Genet. 1998;63:297-304
4. Krieghoff HE, Hofmann TG. Role of nuclear bodies in apoptosis signalling. Biochimica et Biophysica Acta. 2008;1783:2185-94
5. Terris B, Baldin V, Dubois S. et al. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590-7
6. Koken MH, Linares CG, Quignon F. et al. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315-24
7. Valérie LB, Hugues dT. CK2 and PML: Regulating the Regulator. Cell. 2006;126:244-5
8. Grimwade D, Solomon E. Characterisation of the PML/RARa rearrangement associated with t(15;17) acute promyelocytic leukemia. Curr Top Microbiol Immunol. 1997;220:81-112
9. Saurin AJ, Borden KL, Boddy MN. et al. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208-14
10. Soo JY, Seob EJ, Jae HL. A complex,four-way variant t(15;17) in acute promyelocytic leukemia. Cancer Genetics and Cytogenetics. 2006;167:168-71
11. Grisolano JL, Wesselschmidt RL, Pelicci PG. et al. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RARα under control of cathepsin G regulatory sequences. Blood. 1997;89:376-87
12. He LZ, Tribioli C, Riv R. et al. Acute leukemia with promyelocytic features in PML/RARα transgenic mice. Proc Natl Acad Sci USA. 1997;94:5302-7
13. Westervelt P, Lane AA, Pollock JL. et al. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARα expression. Blood. 2003;102:1857-65
14. Early E, Moore MA, Kakizuka A. et al. Transgenic expression of PML/RARα impairs myelopoiesis. Proc Natl Acad Sci USA. 1996;93:7900-4
15. Lane AA, Ley TJ. Neutrophil elastase cleaves PML-RARα and is important for the development of acute promyelocytic leukemia in mice. Cell. 2003;115:305-18
16. Zheng HY, Hu JD. et al. Emodin may induce HL - 60 cell apoptosis througn inhibiting Akt signal pathways. Pharmaceutical journal. 2007;42:1142-6
17. Hayakawa F, Privalsky ML. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell. 2004;5:389-401
18. Shima Y, Shima T, et a1. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol Cell Biol. 2008;28:7126-38
19. Mu ZM, Chin KV, Liu JH. et al. PML a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858-67
20. Ahn MJ, Langenfeld J, Moasser MM. et al. Growth suppression of transformed human bronchial epithelial cells by all-trans-retinoic acid occurs through specific retinoid receptors. Oncogene. 1995;11:2357-64
21. Koken MH, Linares-Cruz G, Quignon F. et al. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315-24
22. Le XF, Yang P, Chang KS. Analysis of the growth and transformation suppressor domains of promyelocytic leukemia gene PML. J Biol Chem. 1996;271:130-5
23. Negorev D, Maul GG. Cellular proteins localized atand interacting within ND10/PML nuclear bodies/PODs suggest functions of a nucleardepot. Oncogene. 2001;20:7234-42
24. Willis S, Catherine LD, Mark GH. et al. The BCL-2-regulated apoptotic pathway. Journal of Cell Science. 2003;116:4053-6
25. Juin P, Geneste O, Raimbaud E. et al. Shooting at survivors: BCL-2 family members as drug targets for cancer. Biochimica et Biophysica Acta. 2004;1644:251-60
Author contact
![]() Corresponding author: Bei-Zhong Liu, Department of Laboratory Medicine, Chongqing Medical University, 1#, Yixueyuan Road, Chongqing, 400016, China. Tel: +86 18716469741, Fax: +86 023-68485006; E-mail: liubeizhongedu.cn.
Corresponding author: Bei-Zhong Liu, Department of Laboratory Medicine, Chongqing Medical University, 1#, Yixueyuan Road, Chongqing, 400016, China. Tel: +86 18716469741, Fax: +86 023-68485006; E-mail: liubeizhongedu.cn.

 Global reach, higher impact
Global reach, higher impact