Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(3):312-319. doi:10.7150/ijms.5455 This issue Cite
Research Paper
Rs143383 in the Growth Differentiation Factor 5 (GDF5) Gene Significantly Associated with Osteoarthritis (OA)-A Comprehensive Meta-analysis
1. Shanghai Key Laboratory for Prevention and Treatment of Bone and Joint Diseases with Integrated Chinese-Western Medicine, Shanghai Institute of Orthopaedics and Traumatology, Department of Orthopaedics, Shanghai Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200025, China;
2. Department of General Surgery, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200025, China;
3. State Key Laboratory of Medical Genomics, Research Center for Experimental Medicine, Shanghai Institute of Hematology, Ruijin Hospital Affiliated to Shanghai Jiaotong University School of Medicine (SJTUSM), Shanghai 200025, China.
* These two authors contributed equally to this work.
Received 2012-10-27; Accepted 2013-1-9; Published 2013-2-5
Abstract
Family, twin, adoption studies show osteoarthritis (OA) has a substantial genetic component. Several studies have shown an association between OA and Growth Differentiation Factor 5 (GDF5), some others have not. Thus, the status of the OA-GDF5 association is uncertain. This meta-analysis was applied to case-control studies of the association between OA and GDF5 to assess the joint evidence for the association, the influence of individual studies, and evidence for publication bias. Relevant studies were identified from the following electronic databases: MEDLINE and current contents before Feb. 2012.
For the case-control studies, the authors found 1) support for the association between OA and GDF5. The rs143383 polymorphism was significantly associated with OA [fixed: OR and 95%CI: 1.193 (1.139-1.249), p<0.001; random: OR and 95%CI: 1.204 (1.135-1.276), p<0.001], 2) no evidence that this association was accounted for by any one study, and 3) no evidence for publication bias. Although the effect size of the association between OA and GDF5 is small, there is suggestive evidence for an association. Further studies are needed to clarify what variant of GDF5 (or some nearby gene) accounts for this association.
Keywords: GDF5, Osteoarthritis, polymorphism, meta-analysis.
Introduction
Primary osteoarthritis (OA) is an idiopathic phenomenon, occurring in previously intact joints, with no apparent initiating factor such as joint injury or developmental abnormalities. It is characterized by the progressive failure of the extracellular cartilage matrix leading to articular cartilage destruction.[1-3] It is widely believed that OA develops from an imbalance between anabolic and catabolic processes or homeostasis of cartilage metabolism.[4-5] Although the detailed aetiology is not currently fully understood, it has been suggested that OA resulted from the combination of aging, hormonal, environmental, and genetic factors.[6-7] It is well known that genetic factors contribute to the susceptibility for OA. Several studies have demonstrated that polymorphisms in many genes might be related to the pathogenesis of OA.[8-18]
Growth differentiation factor 5 (GDF5), also known as cartilage-derived morphogenetic protein 1 or bone morphogenetic protein (BMP) 14, is a member of TGF-beta superfamily.[19] A number of studies have demonstrated that GDF5 plays important roles in musculoskeletal processes, affecting endochondral ossification, synovial joint formation, tendon maintenance, and bone formation.[20-21] Defects of this gene was shown to be correlated to abnormal joint development or skeletal disorders in humans and mice.[22-25] Moreover, it has been reported that the polymorphism in GDF5 gene is related with low expression of the GDF5 protein in knee joint.[26] In addition, GDF5 deficient mice exhibited biomechanical abnormalities in the tendon, which may be associated with altered type I collagen and skeletal abnormalities, one hypothesis of the mechanism behind that was GDF5 might modulate the rate of endochondral bone growth by affecting the duration of the hypertrophic phase in growth plate chondrocytes.[27]
Several studies have shown that the association of rs143383 polymorphism in the promoter region might be a risk factor of OA,[26,28-33] but other groups did not confirm these results.[34] To deal with the ambiguities raised by inconsistent results among molecular genetic studies, Rice suggested that the statistical method of meta-analysis could be used to reconcile conflicting findings.[35] This method is used to examine whether the aggregate evidence across all available studies provides evidence of statistical significance. Thus, to examine the putative association between OA and the GDF5, we applied this comprehensive meta-analysis to all available case-control association studies.
Materials and methods
Search strategy. To assess the total evidence of association between GDF5 gene and OA, we performed this meta-analysis of published studies. We considered all studies that examined the association of the rs143383 polymorphisms with OA. Sources included PUBMED and EMBASE (search last updated in Feb. 2012). The search strategy was based on combinations of the terms ''GDF5'' ''growth and differentiation factor 5'' ''BMP14” ''1104T/C'' ''rs143383'' and ''OA'', and ''osteoarthritis.'' Reference lists in retrieved articles were also screened.
Inclusion and exclusion criteria. All the studies included satisfied all the following criteria: they (1) were association studies between the rs143383 polymorphisms in the GDF5 gene and OA; (2) used disease-free people as controls; (3) provided genotypes or alleles distribution in both case and control groups; (4) were independent studies and the subject groups investigated in each studies did not overlap with other's; (5) were published in peer-reviewed journals and were indexed by PubMed or cited by articles indexed by PubMed. Authors were contacted where clarification was required.
The phenotype definitions were accepted basing on clinical criteria and radiographic criteria. 1) For the clinical criteria, the American College of Rheumatology (ACR; formerly, the American Rheumatism Association) criteria was used [36-38] but also accepted other definitions that may have been preferred by local investigators, if information on ACR criteria was not available. 2) For the radiographic criteria, we used the Kellgren/Lawrence (K/L) classification system (grades 0-4, with 0 representing normal findings and 4 representing severe OA) [39], which is the most widely used scale for identifying and grading OA.[28] A cutoff of K/L grade 2 was used to classify OA, unless the data had been generated with another cutoff and the definition could not be revisited. The phenotype details which were used in every study are shown in Table 1.
Characteristics of the studies included.
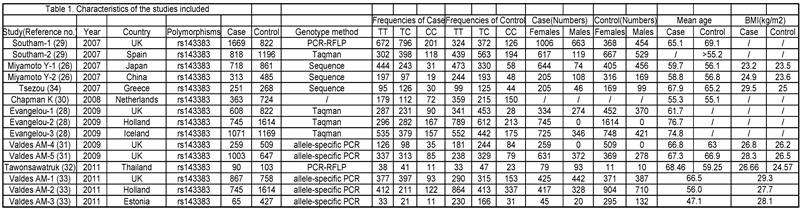
Nonfamilial case-control studies were eligible if the researchers had determined the distribution of genotypes for the polymorphism in OA cases and disease-free controls. We excluded studies with family-based designs in which the analysis was based on linkage considerations.
Only studies published in English were included. Studies presenting non-original data were excluded, such as reviews, editorials, opinion papers, or letters to the editor. Studies using non-human subjects or specimens were excluded. Studies in which rheumatoid, inflammatory, or other forms of arthritis were incorporated in the OA datasets were excluded. Studies with no extractable, numerical data were excluded. Only those articles which had some measure of diagnostic performance were included. Any duplicates which came up in the preliminary search were excluded.
Data extraction. The following data was extracted from all identified studies, by two independent investigators: 1) first author, 2) journal, 3) year of publication, 4) study design, 5) ethnicity of the study population, 6) gender, 7) clinical characteristics, 8) genotyping method, 9) the number of cases and controls or OR and 95%CI (odds ratio and 95 percent confidence interval), 10) country in which the study was conducted and confirmation of diagnosis. Inconsistencies were resolved in consensus.
Statistical analysis. We used a comprehensive meta-analysis to analyze the odds ratios by using the method of Carlin.[40] To determine whether the results of the meta-analysis were unduly influenced by any one study, we recomputed the meta-analysis statistic after deleting each study one at a time. We assessed publication bias by using the method of Egger et al.[41] This method is based on the fact that the precision of the odds ratio increases with larger study groups. The method regresses the standard normal deviate of the odds ratio (the odds ratio divided by its standard error) against the precision of the odds ratio (the inverse of its standard error). In the absence of bias, Egger et al. showed that the regression of the standard normal deviate on precision of the odds ratio should run through the origin (i.e., small study groups with low precision have large standard errors and therefore small standard normal deviates; large study groups have higher precision, smaller standard errors, and large standard normal deviates). The publication bias statistic of Egger et al. is the intercept of the regression, which will be significantly greater than zero in the presence of publication bias.
The effect size was represented by an odds ratio (OR) with 95% confidence interval (CI). Sensitivity analysis was conducted by removing each study and analyzing the others to ensure no single study was totally responsible for overall results. The significance level was set at 0.05, and all P values were two-tailed. Between-study heterogeneity was tested using Cochran's Q statistic, which is considered significant at P < 0.10 [42]. The extent of inconsistency across studies was quantified with the I2 statistic [43]. The I2 ranges between 0 and 100%. For operational purposes, values of 0-24%, 25-50%, 50-75%, and >75% are considered low, moderate, large, and very large, respectively [28,44]. When there was very large or large (>50%) between-study heterogeneity, we used a simulation algorithm to evaluate how many studies had to be removed for the I2 to reach <25% [45].
The comprehensive meta-analysis was performed using Comprehensive Meta-Analysis software (Version 2.2.046, BIOSTAT, Englewood, NJ, USA).
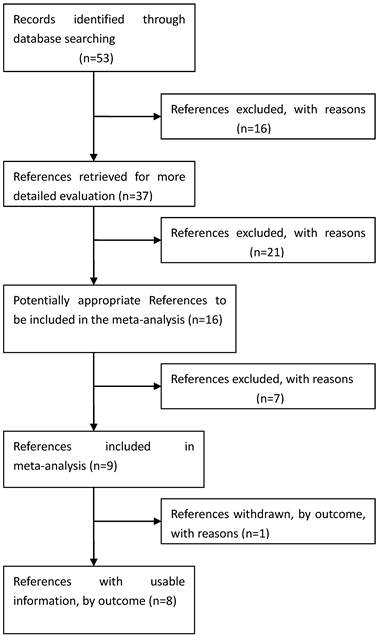
Process used to select published studies for a systematic review and meta-analysis of the relation between GDF5 and OA, 2007-Feb 2012. MeSH, Medical Subject Headings.
Results
Eligible studies. The combined search yielded at least 53 references. After discarding overlapping references and those that clearly did not meet the criteria, 37 references were retained. These references were then filtered to ensure conformity with the inclusion criteria. 21 references were discarded for insufficient and equivocal data (although we tried unsuccessfully to obtain further information from the authors), 7 references were excluded because they were not about the researches of SNPs, and 1 reference was excluded because its data were about lumbar disc degeneration. In total, 15 studies (8 references) met our inclusion criteria, altogether consisting of a total of 7881 cases and 12019 controls.[26, 28-34] The main characteristics of these studies are described in Table 1.
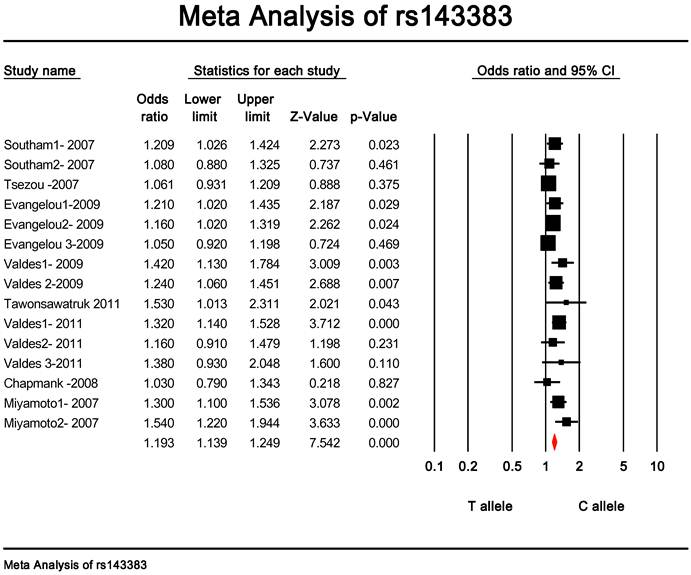
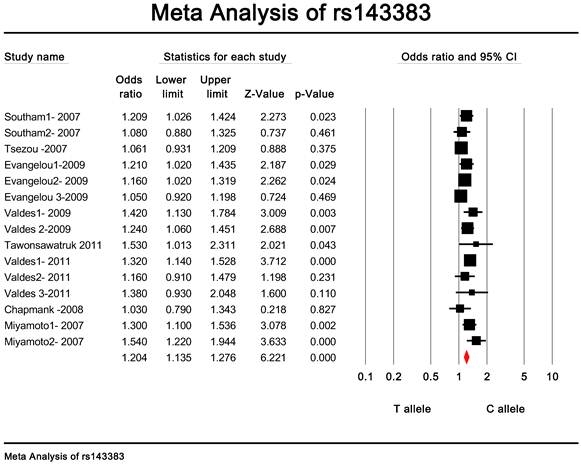
Quantitative data synthesis. The eligible studies for analysis included a total of 7881 cases with OA and 12019 controls (Table 1). The meta-analysis of all the studies about rs143383 polymorphism was significantly associated with OA [fixed: odds ratio and 95 percent confidence interval (CI): 1.193 (1.139-1.249), p<0.001; random: odds ratio and 95 percent confidence interval (CI): 1.204 (1.135-1.276), p<0.001 ] (Fig. 2 and Fig. 3).
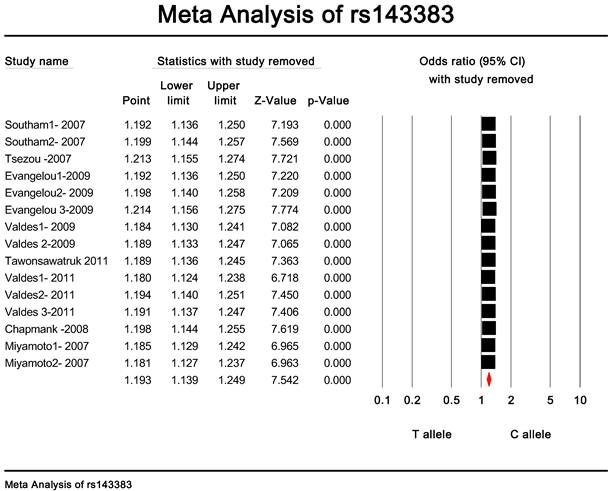
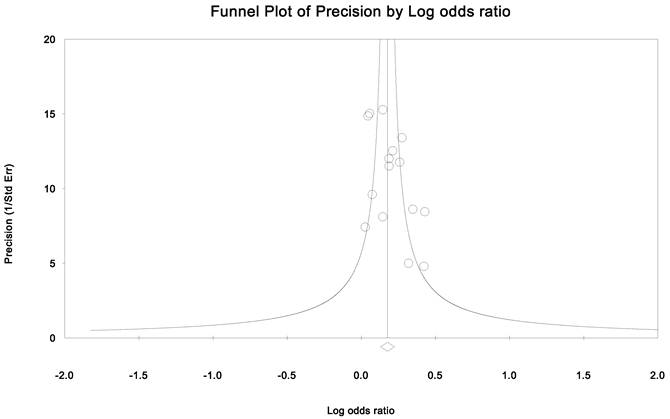
Publication bias and heterogeneity. The sensitivity analysis showed that when any one of the studies was removed, the heterogeneity of the population was not changed deeply. In other words, when one study was removed, the result also showed significant (Fig. 4). This indicated that no heterogeneity existed in the population. There was no evidence that the magnitude of the overall odds ratio estimates changed in the same direction over time. And the Egger's funnel plots of publication bias analysis for the rs143383 polymorphism was shown in Fig. 5.
Meta-analysis of association studies of the rs143383 polymorphism and OA (fixed model). The overall OR is shown. The OR of each study is marked with a black square. The overall OR is indicated by diamond.
Meta-analysis of association studies of the rs143383 polymorphism and OA (random model).The overall OR is shown. The OR of each study is marked with a black square. The overall OR is indicated by diamond.
The sensitivity analysis of rs143383.When any one of the studies was removed, the heterogeneity of the population was not changed.
Egger's funnel plots of publication bias analysis for the rs143383 polymorphism. The larger the deviation from the funnel curve of each study, the more pronounced the asymmetry. Results from small studies will scatter widely at the bottom of the graph, with the spread narrowing among larger studies.
Discussion
The pathogenesis of the development and progression of OA is far from being clear at present. Several studies indicated that variants of the GDF5 gene may contribute to the disease, but the results of genetic association studies were confusing because of the difficulty in replicating significant associations. Different characteristics among studies such as ethnicities, definition of case and control, introduced heterogeneity and made the results of association studies hard to be interpreted. A meta-analysis aiming at finding out the origin of heterogeneity and assessing overall effects of these variants on OA was performed. This comprehensive meta-analysis included data from 8 references including 15 studies with approximately 19900 OA cases and controls. It revealed highly significant evidence of association between the rs143383 polymorphism of GDF5 gene and OA.
GDF5 is an extracellular signaling molecule that participates in bone and cartilage morphogenesis as well as in joint formation.[46] Mutations in human GDF5 gene results in a broad spectrum of skeletal disorders.[47] Based on this functional knowledge, Miyamoto et al. genotyped a number of common GDF5 polymorphisms and demonstrated that rs143383, a T to C transition located in the 5' untranslated region (5'UTR) of the gene, was significantly associated with OA.[26] Further studies have revealed that rs143383 is functional, the OA-associated T-allele mediating reduced GDF5 transcription relative to the C-allele in all of the joint tissues.[47-48] Moreover, the A-allele of -41C/A polymorphism is able to compensate for the reduced expression mediated by the T-allele of rs143383.[49] In some other studies, rs143383 has also been shown to be associate with other phenotypes such as variation in normal height, Achilles tendon pathology, fracture risk and congenital dysplasia of the hip.[50-54] As Loughlin described, this highlights the tendency of a common genotype to influence multiple phenotypes (pleiotropy), and indicates that developmental factors may play an important role on conditions that manifest in the mature individual.[55]
Functional study has demonstrated that T allele of rs143383 was associated with the decrease of GDF5 molecule expression and might increase susceptibility to OA.[26] The differential binding of deformed epidermal autoregulatory factor 1 (DEAF-1) could modulate the expression of GDF5 via this polymorphism.[48] These evidences indicated that the functional polymorphism of GDF5 gene plays critical roles in the etiology of OA. Mouse models have provided a basis for better understanding of the role of GDF5 in skeletogenesis and joint maintenance.[22-24] The brachypodism (bp) mice which carry a functional null allele of GDF5 caused by a frame-shift mutation, exhibited abnormal skeletal and bone development.[56-57] However, Gdf5Bp-J/+ mice appeared phenotypically normal, but does show an increased propensity of developing an OA phenotype when challenged.[58] This model suggested that decreased GDF5 levels in mice contribute to osteoarthritis development, and it is supportive of the genetic data indicating the association between rs143383 polymorphism of GDF5 and human osteoarthritis.
In the present study, we assessed all available literature. This effort to take a comprehensive and even-handed approach to the literature inclusion may have strengthened the robustness of the findings while it avoided publication bias and minimized heterogeneity.[59] Compared with previous study [30], the current meta-analysis pooled larger sample sizes, analyzed them both combined and separately, generated even more significant results with systematic design types and analysis approaches and included tests of heterogeneity, as well as sensitivity analyses. The current results showed that there is a potential association between the rs143383 allele of GDF5 and OA. For greater insight into OA's genetic component, more work is required to confirm the role of other genes that may have a small effect, and to identify new genetic risk factors. The large samples required will necessitate multi-site projects and meta-analyses on the basis of national and international collaboration.
In summary, this meta-analysis supports significant association of marker in the GDF5 gene with OA. It remains unclear why the frequency of the associated alleles varies across studies. Identification of functional variants will probably require biological as well as additional genetic assays.
Acknowledgements
This work is supported by grants from the National Natural Science Foundation of China (31000408); and fund from the National Natural Science Foundation of International Communication and Cooperation Projects (No.81061160510). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributions
J. Liu and H.X. Zhang designed the study and drafted the manuscript. J. Liu, W. Cai, C. He, L.F. Deng and H.X. Zhang performed the statistical analysis. All authors critically revised the manuscript and gave final approval of the article for submission.
Competing Interests
The authors have declared that no competing interests exist.
References
1. Hunter DJ, March L, Sambrook PN. Knee osteoarthritis: the influence of environmental factors. Clin Exp Rheumatol. 2002;20:93-100
2. Peach CA, Carr AJ, Loughlin J. Recent advances in the genetic investigation of osteoarthritis. Trends Mol Med. 2005;11:186-91
3. Dieppe PA, Lohmander LS. Pathogenesis and management of pain in osteoarthritis. Lancet. 2005;365:965-73
4. Felson DT. Osteoarthritis of the knee. N Engl J Med. 2006;354:841-8
5. Sandell LJ, Aigner T. Articular cartilage and changes in arthritis. An introduction: cell biology of osteoarthritis. Arthritis Res. 2001;3:107-13
6. Lanyon P, Muir K, Doherty S. et al. Assessment of a genetic contribution to osteoarthritis of the hip: sibling study. BMJ. 2000;321:1179-83
7. Das SK, Farooqi A. Osteoarthritis. Best Pract Res Clin Rheumatol. 2008;22:657-75
8. Loughlin J, Sinsheimer JS, Mustafa Z. et al. Association analysis of the vitamin D receptor gene, the type I collagen gene COL1A1, and the estrogen receptor gene in idiopathic osteoarthritis. J Rheumatol. 2000;27:779-84
9. Spector TD, MacGregor AJ. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage. 2004;12(Suppl A):39-44
10. Smith AJ, Keen LJ, Billingham MJ. et al. Extended haplotypes and linkage disequilibrium in the IL1R1-IL1A-IL1B-IL1RN gene cluster: association with knee osteoarthritis. Genes Immun. 2004;5:451-60
11. Aigner T, Dudhia J. Genomics of osteoarthritis. Curr Opin Rheumatol. 2003;15:634-40
12. Kizawa H, Kou I, Iida A. et al. An aspartic acid repeat polymorphism in aspirin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat Genet. 2005;37:138-44
13. Mabuchi A, Ikeda T, Fukuda A. et al. Identification of sequence polymorphisms of the COMP (cartilage oligomeric matrix protein) gene and association study in osteoarthrosis of the knee and hip joints. J Hum Genet. 2001;46:456-62
14. Mototani H, Mabuchi A, Saito S. et al. A functional single nucleotide polymorphism in the core promoter region of CALM1 is associated with hip osteoarthritis in Japanese. Hum Mol Genet. 2005;14:1009-17
15. Ikeda T, Mabuchi A, Fukuda A. et al. Identification of sequence polymorphisms in two sulfation-related genes, PAPSS2 and SLC26A2, and an association analysis with knee osteoarthritis. J Hum Genet. 2001;46:538-43
16. Jiang Q, Shi D, Yi L. et al. Replication of the association of the aspartic acid repeat polymorphism in the asporin gene with knee-osteoarthritis susceptibility in Han Chinese. J Hum Genet. 2006;51:1068-72
17. Valdes AM, Spector TD. The contribution of genes to osteoarthritis. Rheum Dis Clin North Am. 2008;34:581-603
18. Valdes AM, Loughlin J, Timms KM. et al. Genome-wide association scan identifies aprostaglandinendoperoxide synthase 2 variant involved in risk of knee osteoarthritis. Am J Hum Genet. 2008;82:1231-4
19. Buxton P, Edwards C, Archer CW. et al. Growth/differentiation factor-5 (GDF-5) and skeletal development. J Bone Joint Surg Am. 2001;83(Suppl 1):23- 30
20. Nishitoh H, Ichijo H, Kimura M. Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor-5. J Biol Chem. 1996;271:21345-52
21. Mikic B, Battaglia TC, Taylor EA. The effect of growth/differentiation factor-5 deficiency on femoral composition and mechanical behavior in mice. Bone. 2002;30:733-7
22. Masuya H, Nishida K, Furuichi T. et al. A novel dominant-negative mutation in Gdf5 generated by ENU mutagenesis impairs joint formation and causes osteoarthritis in mice. Hum Mol Genet. 2007;16:2366-75
23. Chhabra A, Tsou D, Clark RT. et al. GDF-5 deficiency in mice delays Achilles tendon healing. J Orthop Res. 2003;21:826-35
24. Harada M, Takahara M, Zhe P. et al. Developmental failure of the intra-articular ligaments in mice with absence of growth differentiation factor 5. Osteoarthritis Cartilage. 2007;15:468-74
25. Nickel J, Kotzsch A, Sebald W. A single residue of GDF-5 defines binding specificity to BMP receptor IB. J Mol Biol. 2005;349:933-47
26. Miyamoto Y, Mabuchi A, Shi D. et al. A functional polymorphism in the 5'-UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529-33
27. Mikic B, Clark RT, Battaglia TC. Altered hypertrophic chondrocyte kinetics in GDF-5 deficient murine tibial growth plates. J Orthop Res. 2004;22:552-6
28. Evangelou E, Chapman K, Meulenbelt I. Large-scale analysis of association between GDF5 (rs143383) and FRZB (rs7775 and rs288326) variants and hip, knee and hand osteoarthritis. Arthritis Rheum. 2009;60:1710-21
29. Southam L, Rodriguez-Lopez J, Wilkins JM. et al. An SNP in the 5'-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and within vivo differences in allelic expression in articular cartilage. Hum Mol Genet. 2007;16:2226-32
30. Chapman K, Takahashi A, Meulenbelt I. et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5' UTR of GDF5 with osteoarthritis susceptibility. Hum Mol Genet. 2008;17:1497-504
31. Valdes AM, Spector TD, Doherty S. et al. Association of the DVWA and GDF5 polymorphisms with osteoarthritis in UK populations. Ann Rheum Dis. 2009;68:1916-20
32. Tawonsawatruk T, Changthong T, Pingsuthiwong S. et al. A genetic association study between growth differentiation factor 5 (GDF 5) polymorphism and knee osteoarthritis in Thai population. J Orthop Surg Res. 2011;6:47
33. Valdes AM, Evangelou E, Kerkhof HJ. et al. The GDF5 rs143383 polymorphism is associated with osteoarthritis of the knee with genome-wide statistical significance. Ann Rheum Dis. 2011;70:873-5
34. Tsezou A, Satra M, Oikonomou P. et al. The growth differentiation factor 5 (GDF5) core promoter polymorphism is not associated with knee osteoarthritis in the Greek population. J Orthop Res. 2008;26:136-40
35. Rice JP. The role of meta-analysis in linkage studies of complex traits. Am J Med Genet. 1997;74:112-4
36. Altman R, Alarcon G, Appelrouth D. et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the hand. Arthritis Rheum. 1990;33:1601-10
37. Altman R, Alarcon G, Appelrouth D. et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the hip. Arthritis Rheum. 1991;34:505-14
38. Altman R, Asch E, Bloch D. et al. Development of criteria for the classification and reporting of osteoarthritis: classification of osteoarthritis of the knee. Arthritis Rheum. 1986;29:1039-49
39. Kellgren JH, Lawrence JS. Radiological assessment of osteoarthrosis. Ann Rheum Dis. 1957;16:494-502
40. Carlin JB. Meta-analysis for 2 × 2 tables: a Bayesian approach. Stat Med. 1992;11:141-58
41. Egger M, Davey Smith G, Schneider M. et al. Bias in metaanalysis detected by a simple, graphical test. Br Med J. 1997;315:629-34
42. Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1-14
43. Higgins JP, Thompson SG. Quantifying heterogeneity in a metaanalysis. Stat Med. 2002;21:1539-58
44. Higgins JP, Thompson SG, Deeks JJ. et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-60
45. Patsopoulos NA, Evangelou E, Ioannidis JP. Sensitivity of between- study heterogeneity in meta-analysis: proposed metrics and empirical evaluation. Int J Epidemiol. 2008;37:1148-57
46. Francis-West PH, Abdelfattah A, Chen P. et al. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305-15
47. Byrnes AM, Racacho L, Nikkel SM. et al. Mutations in GDF5 presenting as semidominant brachydactyly A1. Hum Mutat. 2010;31:1155-62
48. Egli R, Southam L, Wilkins JM. et al. Functional analysis of the osteoarthritis susceptibility-associated GDF5 regulatory polymorphism. Arthritis Rheum. 2009;60:2055-64
49. Dodd AW, Syddall CM, Loughlin J. A rare variant in the osteoarthritis-associated locus GDF5 is functional and reveals a site that can be manipulated to modulate GDF5 expression. Eur J Hum Genet. 2012 [Epub ahead of print]
50. Sanna S, Jackson AU, Nagaraja R. et al. Common variants in the GDF5eUGCC region are associated with variation in human height. Nat Genet. 2008;40:198-203
51. Posthumus M, Collins M, Cook J. et al. Components of the transforming growth factor-b family and the pathogenesis of human Achilles tendon pathology e a genetic association study. Rheumatology (Oxford). 2010;49:2090-7
52. Vaes RB, Rivadeneira F, Kerkhof JM. et al. Genetic variation in the GDF5 region is associated with osteoarthritis, height, hip axis length and fracture risk: the Rotterdam study. Ann Rheum Dis. 2009;68:1754-60
53. Dai J, Shi D, Zhu P. et al. Association of a single nucleotide polymorphism in growth differentiate factor 5 with congenital dysplasia of the hip: a case-control study. Arthritis Res Ther. 2008;10:R126
54. Rouault K, Scotet V, Autret S. et al. Evidence of association between GDF5 polymorphisms and congenital dislocation of the hip in a Caucasian population. Osteoarthritis Cartilage. 2010;18:1144-9
55. Loughlin J. Genetic indicators and susceptibility to osteoarthritis. Br J Sports Med. 2011;45:278-82
56. Storm EE, Huynh TV, Copeland NG. et al. Limb alterations in brachypodism mice due to mutations in a new member of the TGFb-superfamily. Nature. 1994;368:639-43
57. Takahara M, Harada M, Guan D. et al. Developmental failure of phalanges in the absence of growth/differentiation factor 5. Bone. 2004;35:1069-76
58. Daans M, Luyten FP, Lories RJ. GDF5 deficiency in mice is associated with instability-driven joint damage, gait and subchondral bone changes. Ann Rheum Dis. 2011;70:208-13
59. Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439-44
Author contact
![]() Corresponding author: 197 Ruijin er Road, 200025, Shanghai, People's Republic of China. Tel: +86 13621722453 e-mail: lianfudengcom (LF. Deng).
Corresponding author: 197 Ruijin er Road, 200025, Shanghai, People's Republic of China. Tel: +86 13621722453 e-mail: lianfudengcom (LF. Deng).

 Global reach, higher impact
Global reach, higher impact