Impact Factor
ISSN: 1449-1907
Int J Med Sci 2012; 9(10):901-908. doi:10.7150/ijms.4655 This issue Cite
Research Paper
Quantification of BCR-ABL mRNA in Plasma/Serum of Patients with Chronic Myelogenous Leukemia
1. Laboratory of Hematology and Oncology, Graduate School of Health Sciences, Niigata University, 1-746 Asahimachi-dori, Chuo-ku, Niigata, 951-8518 Japan;
2. Division of Bone Marrow Transplantation, Niigata University Medical and Dental Hospital, 1-754 Asahimachi-dori, Chuo-ku, Niigata, 951-8520, Japan;
3. Division of Hematology, Endocrinology and Metabolism, Department of Internal Medicine, Faculty of Medicine, Niigata University, Niigata, 951-8510, Japan.
Received 2012-5-26; Accepted 2012-10-30; Published 2012-11-12
Abstract
Quantification of tumor-associated mRNA extracted from blood cells/tissues containing tumor cells is used for evaluation of treatment efficacy or residual tumor cell burden in tumors including leukemia. However, this method using tumor cell-containing blood/tissue is difficult to evaluate the whole tumor cell burden in the body. In order to establish an efficient method to evaluate the whole tumor cell burden in the body, we tried to quantify tumor-associated mRNA existing in plasma/serum instead of leukemia cell-containing blood cells in patients with chronic myelogenous leukemia (CML) and compared the levels of BCR-ABL mRNA between plasma/serum and peripheral blood cells. mRNA of BCR-ABL, WT1 or GAPDH (control molecule) was detected by real-time RT-PCR using RNA extracted from plasma/serum of almost all the patients with CML. Copy numbers of BCR-ABL mRNA were significantly correlated between plasma/serum and peripheral blood cells. However, levels of BCR-ABL mRNA extracted from serum were low compared with those extracted with peripheral blood cells. The present findings suggest that although real-time RT-PCR of mRNA existing in plasma/serum could be used for evaluating the whole tumor cell burden in the body, it's required to establish an efficient method to quantify plasma/serum mRNA by nature without degrading during the procedure.
Keywords: serum, plasma, mRNA, CML, BCR-ABL, WT1, real-time RT-PCR.
Introduction
Real-time quantitative reverse transcriptase-polymerase chain reaction (real-time RT-PCR) of tumor-associated mRNA is useful and routinely used for the diagnosis of tumors possessing characteristic molecular manifestations. In addition, real-time RT-PCR is important for the evaluation of the therapy effectiveness in patients with various leukemia expressing specific gene rearrangements (such as BCR-ABL, PML-RARα, AML1-ETO) or nonspecific tumor markers such as WT1. In hematological malignancies such as leukemia, leukemia-associated mRNA is extracted from peripheral blood (PB) cells or bone marrow (BM) cells, which are infiltrated with leukemia cells. In the diagnosis of tumor, tumor cell-containing tissue is ideal source for extracting tumor-associated mRNA. However, in the evaluation of whole body residual leukemia cells after treatment with anti-leukemic drugs (like residual CML cells after imatinib therapy), mRNA extracted from PB or BM cells might not be appropriate, since leukemia cells are not distributed evenly between PB and BM or among BMs at different sites in patients. Therefore, the quantification of mRNA obtained from cell samples of leukemia patients by using real-time RT-PCR could not always reflect the total burden of leukemia cells in whole body. Moreover, in case of solid tumor, it might be difficult to obtain tissues containing tumor cells unless operative procedure is undertaken.
Recently leukemia cells were demonstrated to pour their RNA into surrounding bone marrow fluids or peripheral blood [1]. Although RNase is present in peripheral blood, free mRNA was demonstrated to exist in blood [2, 3]. We speculated that plasma/serum tumor-associated mRNA reflects the whole burden of leukemia cells in the entire body of patient and plasma/serum represents a reliable source of mRNA for quantification of copy number of tumor-associated mRNA by using real-time RT-PCR. In the present study, we explored the significance of quantifying BCR-ABL or WT1 mRNA present in plasma/serum of the patients with CML for evaluating whole CML cells in the body.
Materials and Methods
Specimens from CML Patients
Plasma, serum and cell samples were collected from patients with CML and healthy volunteers, according to an Institutional review board-approved protocol. All patients provided written informed consents. The diagnosis of CML was made by the detection of both Ph chromosome by karyotype study and BCR-ABL gene rearrangement by FISH analysis.
mRNA Extraction
Total RNA was extracted from 1 ml plasma/serum using the NucliSens extraction kit (magnetic silica method) (BioMerieux, Boxtel, The Netherlands) or Trizol Reagent (Invitrogen, Carlsbad, CA, USA). In some patients, plasma/serum were carefully prepared not to include cells by repeated centrifugation and using a millipore filter. Total RNA was extracted also from peripheral blood cell or bone marrow cell samples. In some patients with CML, plasma/serum and cells were collected simultaneously and mRNA extracted from both sample sources were compared for the quantification of BCR-ABL mRNA. Most plasma/serum were cryopreserved in -80°C and thawed for mRNA extraction. All total RNA extracted from 1 ml plasma/serum were diluted with 50 μl of distilled water.
Real-time RT-PCR
4.75 μl out of 50 μl total mRNA (extracted from 1 ml plasma/serum) were reverse-transcribed into cDNA in a 10 μl total volume using random hexamer primers and avian myeloblastosis virus (AMV) reverse transcriptase according to the manufacturer's protocol (Takara, Kyoto, Japan). 5 μl out of 10 μl reverse-transcribed solution mixture were amplified by using real-time quantitative PCR of 20 μl reaction volume including primers for Major BCR-ABL (b2a2/b3a2), WT1 and GAPDH. PCR was performed in 3 different methods, which were two kinds of cyber green methods and a fluorescence resonance energy transfer hybridization (Universal probe) method. In cyber green method using LightCycler-FastStart DNA master SYBR Green I reagents with LightCycler (Roche diagnostics, Branchburg, NJ, USA), PCR conditions were 45 cycles of denaturing (95°C for 10 sec), annealing (60°C for 10 sec) and extension (72°C for 20 sec). In the method using SYBR Premix ExTaq reagents with Thermal Cycler Dyce (Takara), PCR conditions were 40 cycles of denaturing (95°C for 10 sec) and annealing (60°C for 30 sec).. In fluorescence resonance energy transfer hybridization method using FastStart Universal Probe Master (Roche diagnostics) with LightCycler, PCR conditions were 45 cycles of denaturing (95°C for 10 sec) and annealing (60°C for 25 sec). To control for both the integrity of the sample and any inter-sample variation in the preparation of mRNA, the reference gene GAPDH was amplified under the identical reaction condition in a different tube.
The primers used for both cyber green methods were as follows. BCR-ABL upper primer: 5'-GGAGCAGCAGAAGAAGTGTTT-3', BCR-ABL lower primer: 5'-TGGGTCCAGCGAGAAGGTTTT-3', BCR-ABL product size: b3a2; 293bp, b2a2; 218bp (for LightCycler-FastStart DNA master SYBR Green I reagents with LightCycler), BCR-ABL upper primer: 5'-GCATTCCGCTGACCATCAATAAG-3', BCR-ABL lower primer: 5'-CCTGAGGCTCAAAGTCAGATGCTAC-3', BCR-ABL product size: 144 bp (for SYBR Premix ExTaq reagents with Thermal Cycler Dyce), WT1 upper primer: 5'-AGCACAGGGTACGAGAGCGATAAC-3', WT1 lower primer: 5'-TATTGCAGCCTGGGTAAGCACA-3', WT1 product size: 193 bp for b3a2 (for both LightCycler-FastStart DNA master SYBR Green I reagents with LightCycler and SYBR Premix ExTaq reagents with Thermal Cycler Dyce), GAPDH upper primer: 5'-GCACCGTCAAGGCTGAGAAC-3', GAPDH lower primer: 5'-TGGTGAAGACGCCAGTGGA-3', GAPDH product size: 138 bp (for both LightCycler-FastStart DNA master SYBR Green I reagents with LightCycler and SYBR Premix ExTaq reagents with Thermal Cycler Dyce).
The primers used for Universal probe method were as follows. BCR-ABL upper primer: 5'-GTCCACTCAGCCACTGGATT-3', BCR-ABL lower primer: 5'-TGAGGCTCAAAGTCAGATGC-3', BCR-ABL product size: 74 bp for b3a2 (Universal ProbeLibrary probe: #161 (cat.no.04694481001).
Measurement of copy number of mRNA amplified from 1 ml plasma/serum
Weight (μg) of mRNA amplified from 1 ml of plasma/serum was calculated as follows.
mRNA amplified from 1 ml plasma/serum (μg) = weight of amplified DNA (pg: calculated using standard curve depicted according to cycle number and log concentration of real-time PCR products of plasmid containing BCR-ABL/WT1/GAPDH) x 50 (μl: dilution volume of RNA extracted from plasma/serum) / 4.75 (μl: volume of diluted RNA solution processed to RT) x 10 (μl: total volume of RNA mixture for RT) / 5 (μl: volume of RT product solution processed to PCR) x 10-6
Thereafter, copy number of mRNA amplified from 1 ml plasma/serum was calculated as follows.
Copy number of mRNA amplified from 1 ml plasma/serum = weight of mRNA amplified from 1 ml plasma/serum (μg) x 10-6 (g/μg) / 660 (g/mol) x 1/N x (6.022 x 1023)
660: average molecular weight of nucleic acid pair (dsDNA)
N: number of nucleic acid pair in each DNA molecule (BCR-ABL; 1500 bp, WT1; 3030 bp)
6.022 x 1023 molecules: 1 mol
Statistical analysis
Correlation between copy numbers of BCR-ABL mRNA in paired serum and peripheral blood cell samples were performed using Spearman's correlation coefficients in GraphPad Prism (GraphPad Software La Jolla, CA, USA).
Results
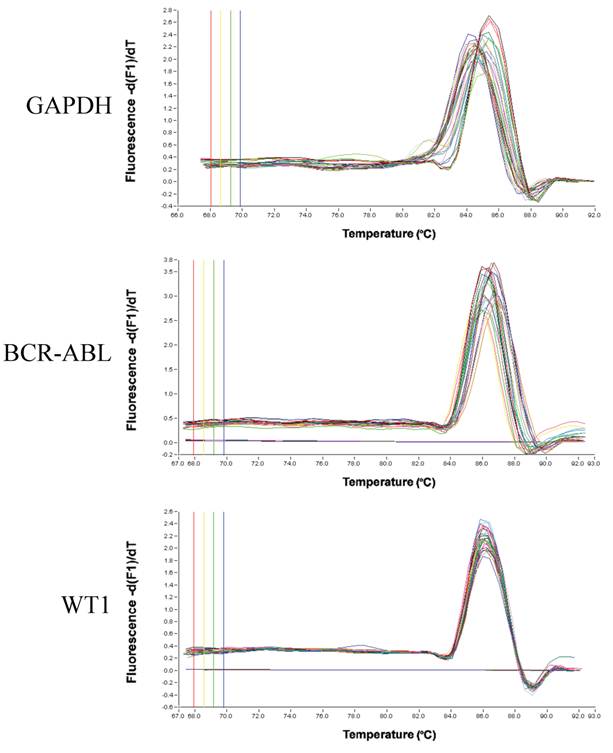
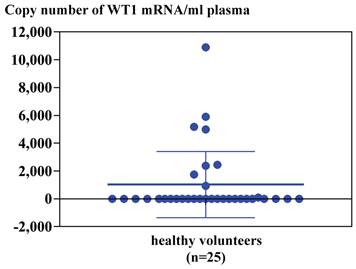
Real-time RT-PCR for GAPDH, BCR-ABL or WT1 mRNA extracted from plasma or serum of CML patients using LightCycler-FastStart DNA master SYBR Green I reagents with LightCycler. Melting temperature (Tm) peak curves demonstrated the isoforms for GAPDH, two peaks for b3a2 and b2a2 of BCR-ABL gene rearrangement and one peak for WT1 (Figure 1). As to RNA extracted from plasma/serum of healthy volunteers by using a NucliSens extraction kit or Trizol Reagent, GAPDH mRNA was detected by real-time RT-PCR in six samples out of 25 (24%) in plasma and five samples out of 10 (50 %) in serum. Although WT1 mRNA was detected in eight samples out of 25 (32%) in plasma and two samples out of 10 (20 %) in serum of healthy volunteers, BCR-ABL mRNA was not detected in both normal plasma and serum (Table 1). Copy number of WT1 RNA in 1 ml plasma/serum of healthy volunteers was 1,000-11,000 copies in plasma/serum samples from which WT1 mRNA was demonstrated to be present (Figure 2).
Real-time RT-PCR of RNA extracted from plasma/serum of healthy volunteers
| Type of samples | number of samples | GAPDH | BCR-ABL | WT1 | |||
|---|---|---|---|---|---|---|---|
| + | (%) | + | (%) | + | (%) | ||
| plasma | 25 | 6 | (24) | 0 | (24) | 8 | (32) |
| serum | 10 | 5 | (50) | 0 | (50) | 2 | (20) |
| total | 35 | 11 | (31) | 0 | (31) | 10 | (29) |
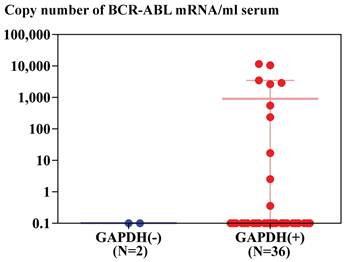
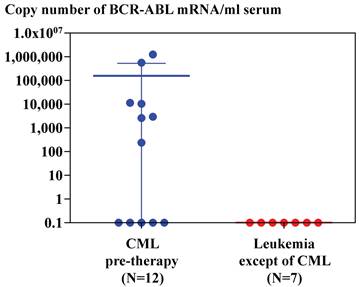
Real-time RT-PCR of GAPDH and BCR-ABL using RNA extracted from serum of patients with CML by magnetic silica method demonstrated that GAPDH mRNA was detected in thirty six samples out of 38 patients with CML (95%) and BCR-ABL mRNA was detected in ten samples out of 36 patients with CML (28%), in whom GAPDH mRNA was detected. BCR-ABL mRNA was not detected in two patients with CML, in whom GAPDH mRNA was not detected (Figure 3). BCR-ABL mRNA was detected in serum or plasma of seven out of 12 patients with CML (60%), who had not been treated with anti-leukemic drug, but BCR-ABL mRNA was not detected in serum or plasma of seven patients, whose diagnosis was not CML (Figure 4).
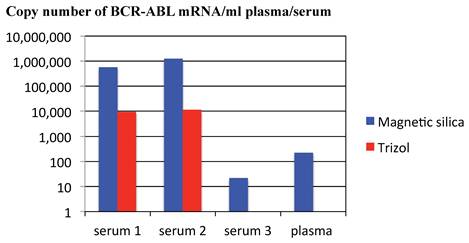
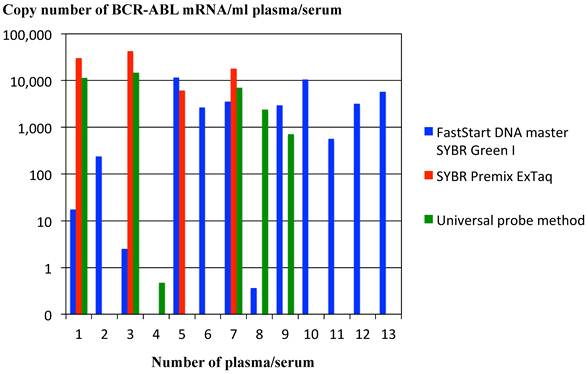
Efficiency for extracting RNA from plasma/serum of patients with CML was compared between magnetic silica method and Trizol reagent method. Real-time RT-PCR of BCR-ABL mRNA extracted from three sera and one plasma of CML patients demonstrated that BCR-ABL mRNA was detected in all of three sera and one plasma in case magnetic silica method was used and BCR-ABL mRNA was not detected in one serum and one plasma in case Trizol reagent method was used for RNA extraction. Copy number of BCR-ABL mRNA was higher in RNA extracted by magnetic silica method compared with that extracted by Trizol reagent method in the sera, in which BCR-ABL mRNA was detected in both extracting method of magnetic silica method and Trizol reagent method. These findings suggested that magnetic silica method is superior to Trizol reagent method in extracting RNA from plasma/serum (Figure 5). Efficiency for real-time RT-PCR of RNA extracted from plasma/serum of patients with CML was compared between two procedures of SYBR Green I (LightCycler-FastStart DNA master SYBR Green I reagents & SYBR Premix ExTaq reagents) and Universal probe method. BCR-ABL mRNA was detected in twelve out of 13 samples of CML patients in real-time RT-PCR using LightCycler-FastStart DNA master SYBR Green I reagents method, in four out of 13 samples in real-time RT-PCR using SYBR Premix ExTaq reagents, and in 6 out of 13 samples in real-time RT-PCR using Universal probe method. These data showed that sensitivity for detecting mRNA in plasma/serum of the patients with tumor varies from method to method of real-time RT-PCR (Figure 6).
Melting temperature (Tm) peak curves of real-time RT-PCR product for GAPDH, BCR-ABL or WT1 mRNA extracted from plasma or serum of CML patients. PCR was performed by cyber green method using LightCycler-FastStart DNA master SYBR Green I reagents with LightCycler.
Copy number of WT1 mRNA amplified from 1 ml plasma of healthy volunteers (n=25). Copy numbers were calculated by standard curves depicted by real-time PCR of plasmid containing WT1 cDNA as described in Materials and Method.
Copy number of BCR-ABL mRNA amplified from mRNA extracted from 1 ml serum of patients with CML using silica gel method. Copy numbers were calculated by standard curves depicted by real-time PCR of plasmid containing BCR-ABL cDNA as described in Materials and Method. Samples were divided into tow groups, depending on the detection of GAPDH mRNA in serum (36 samples) or not (2 samples).
Copy number of BCR-ABL mRNA amplified from mRNA extracted from 1 ml serum of patients with CML (n=12) and leukemia patients except of CML (n=7) using silica gel method. Copy numbers were calculated by standard curves depicted by real-time PCR of plasmid containing BCR-ABL cDNA as described in Materials and Method. Average and SD of copy numbers of BCR-ABL mRNA in CML patients is 154,295 ± 381,878.
Comparison of methods for extracting RNA from plasma/serum of patients with CML. Copy number of BCR-ABL mRNA was compared between RNA extracted by magnetic silica method and that extracted by Trizol reagent method. The graph is a representative data among three independent experiments, the other two of which were performed at different time by using different samples (plasma or serum). The other series of experiments demonstrated the similar results to the representative data shown in the Figure.
Comparison of methods for real-time PCR of cDNA reverse-transcribed from RNA extracted from plasma/serum of patients with CML. Copy number of BCR-ABL mRNA was compared among real-time PCR using LightCycler-FastStart DNA master SYBR Green I reagents, PCR using SYBR Premix ExTaq reagents and PCR using Universal probe method. The graph is a representative data among three independent experiments, the other two of which were performed at different time by using different samples (plasma or serum). The other series of experiments demonstrated the similar results to the representative data shown in the Figure.
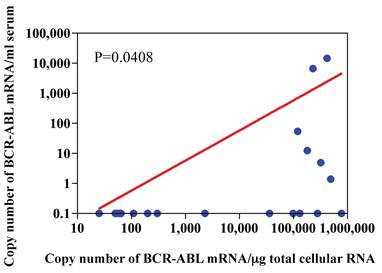
Number of copies of BCR-ABL mRNA were compared between RNA extracted from 1 ml of serum and that from 1 μg of total cellular RNA extracted from peripheral blood cells (including leukemia cells) of the patients with CML. Copy numbers of BCR-ABL mRNA from 1 ml of serum was significantly correlated with those of BCR-ABL mRNA from 1 μg of total cellular RNA of patients with CML (p=0.048, n=18). However, copy numbers of BCR-ABL mRNA from 1 ml of serum were low compared with those of BCR-ABL mRNA from 1 μg of total cellular RNA of the patients with CML (Figure 7).
Relationship between copy number of BCR-ABL mRNA in μg total cellular RNA and that in 1ml serum. Peripheral blood cells and serum were drawn simultaneously from patients with CML, and RNA was extracted from peripheral blood cells and serum using silica gel method. Real-time PCR was undertaken by using LightCycler-FastStart DNA master SYBR Green I reagents.
Discussion
Possible presence of nucleic acid in human plasma was first reported in 1970s [4, 5]. However, the other report said that it's impossible for plasma RNA to exist for long period because of the presence of RNase in plasma [6]. In 1999 Kopreski et al reported that human mRNA can be extracted and amplified from serum and suggested tumor mRNA amplified from serum could be an important tool in cancer diagnostics and monitoring [7]. Thereafter, different circulating cell-free nucleic acids such as DNA, mRNA and microRNA have been detected in the plasma/serum of various cancer patients [8-11]. There are some reports showing plasma RNA is protected from RNase digestion by particle-association [12] such as apoptotic body [13]. Plasma/serum RNA has been shown to be resistant to freeze/thaw cycle [14].
Some investigators have suggested that tumor mRNA in plasma is sensitive to differentiate between malignant and nonmalignant nature and may be a useful marker for early tumor diagnosis [15]. Moreover, the elevated concentration of circulating RNA has been shown to be closely associated with poor prognosis in esophageal cancer, breast cancer or lymphoma [17-19]. The increase of circulating cell-free hTERT mRNA was demonstrated to be correlated with reduced disease-free survival and overall survival in gastric cancer patients by Kang et al. [20]. As to plasma mRNA of CML-derived genes, Ma et al have reported that plasma was a reliable source for monitoring BCR-ABL mRNA levels and that minimal residual disease detection from plasma was more sensitive than from cell samples. They suggested that absolute levels of BCR-ABL mRNA per unit volume of plasma may reflect tumor load [1].
The present study also demonstrated that mRNA could be detected in cryopreserved plasma/serum by real-time RT-PCR in both normal persons and CML patients. Real-time RT-PCR of house keeping gene (GAPDH) in plasma revealed that mRNA is more frequent to be detected in CML patients compared with healthy volunteers, which suggested that a higher amount of mRNA is circulating in CML patients than healthy volunteers. BCR-ABL mRNA as well as WT1 mRNA could be amplified by real-time RT-PCR in patients with CML and copy number of mRNA was significantly correlated between plasma-derived and cell-derived mRNA. However, the present study showed that copy numbers of tumor-associated mRNA are absolutely low in plasma/serum compared with those of RNA extracted from peripheral blood cells containing leukemia cells. We compared the methods of extracting RNA from plasma/serum and the kinds of real-time RT-PCR of extracted RNA, which suggested that copy numbers of mRNA and the frequency of detecting mRNA in plasma/serum varied from method to method. There is a report describing that RNA is stable in serum for up to only 3 hours but is destroyed immediately by addition of detergents and that detection of tumor-associated mRNA in serum from cancer patients and controls was infrequent and inconsistent [3]. We conclude that it's definite that there exists tumor-associated mRNA in plasma/serum from tumor patients and copy numbers of plasma/serum mRNA reflect the burden of whole tumor cells in the body. However, in order to detect plasma/serum tumor-associated mRNA by nature, it's required to investigate the biology of plasma/serum mRNA and to evaluate the proper methods for preserving plasma/serum, extracting mRNA and amplifying cDNA of interest.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Ma W, Tseng R, Gorre M. et al. Plasma RNA as an alternative to cells for monitoring molecular response in patients with chronic myeloid leukemia. Haematologica. 2007;92:170-5
2. Silva JM, Dominguez G, Silva J. et al. Detection of epithelial messenger RNA in the plasma of breast cancer patients is associated with poor prognosis tumor characteristics. Clinical cancer research: an official journal of the American Association for Cancer Research. 2001;7:2821-5
3. El-Hefnawy T, Raja S, Kelly L. et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clinical chemistry. 2004;50:564-73
4. Kamm RC, Smith AG. Nucleic acid concentrations in normal human plasma. Clinical chemistry. 1972;18:519-22
5. Stroun M, Anker P, Maurice P. et al. Circulating nucleic acids in higher organisms. International review of cytology. 1977;51:1-48
6. Kamm RC, Smith AG. Ribonuclease activity in human plasma. Clinical biochemistry. 1972;5:198-200
7. Kopreski MS, Benko FA, Kwak LW. et al. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 1999;5:1961-5
8. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature reviews Cancer. 2011;11:426-37
9. Lo KW, Lo YM, Leung SF. et al. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clinical chemistry. 1999;45:1292-4
10. Chen XQ, Bonnefoi H, Pelte MF. et al. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2000;6:3823-6
11. Kopreski MS, Benko FA, Gocke CD. Circulating RNA as a tumor marker: detection of 5T4 mRNA in breast and lung cancer patient serum. Annals of the New York Academy of Sciences. 2001;945:172-8
12. Ng EK, Tsui NB, Lam NY. et al. Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals. Clinical chemistry. 2002;48:1212-7
13. Hasselmann DO, Rappl G, Tilgen W. et al. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clinical chemistry. 2001;47:1488-9
14. Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clinical chemistry. 2002;48:1647-53
15. Dasi F, Martinez-Rodes P, March JA. et al. Real-time quantification of human telomerase reverse transcriptase mRNA in the plasma of patients with prostate cancer. Annals of the New York Academy of Sciences. 2006;1075:204-10
16. Kudo Y, Ochi T, Shimada H. et al. Utility of plasma circulating mRNA as a marker to detect hepatic injury. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 2008;70:993-5
17. Takahashi S, Miura N, Harada T. et al. Prognostic impact of clinical course-specific mRNA expression profiles in the serum of perioperative patients with esophageal cancer in the ICU: a case control study. Journal of translational medicine. 2010;8:103
18. Garcia V, Garcia JM, Pena C. et al. Free circulating mRNA in plasma from breast cancer patients and clinical outcome. Cancer letters. 2008;263:312-20
19. Garcia V, Garcia JM, Silva J. et al. Extracellular tumor-related mRNA in plasma of lymphoma patients and survival implications. PloS one. 2009;4:e8173
20. Kang Y, Zhang J, Sun P. et al. Circulating cell-free human telomerase reverse transcriptase mRNA in plasma and its potential diagnostic and prognostic value for gastric cancer. International journal of clinical oncology / Japan Society of Clinical Oncology. 2012
21. Ma W, Jilani I, Gorre M. et al. Plasma as a source of mRNA for determining IgV(H) mutation status in patients with chronic lymphocytic leukaemia. Br J Haematol. 2006;133:690-2
Author contact
![]() Corresponding author: Miwako Narita. Laboratory of Hematology and Oncology, Graduate School of Health Sciences, Niigata University, 2-746 Asahimachi-dori, Chuo-ku, Niigata, 951-8518 Japan. Tel.: +81 25227 0836 Fax: +81 25227 0836 E-mail: naritaminiigata-u.ac.jp.
Corresponding author: Miwako Narita. Laboratory of Hematology and Oncology, Graduate School of Health Sciences, Niigata University, 2-746 Asahimachi-dori, Chuo-ku, Niigata, 951-8518 Japan. Tel.: +81 25227 0836 Fax: +81 25227 0836 E-mail: naritaminiigata-u.ac.jp.

 Global reach, higher impact
Global reach, higher impact