Impact Factor
ISSN: 1449-1907
Int J Med Sci 2006; 3(2):35-40. doi:10.7150/ijms.3.35 This issue Cite
Review
Hepatitis C Virus Serologic and Virologic Tests and Clinical Diagnosis of HCV-Related Liver Disease
Department of Virology, INSERM U635, Henri Mondor Hospital, University of Paris XII, Créteil, France
Received 2006-1-12; Accepted 2006-3-15; Published 2006-4-1
Abstract
The use of serological and virological tests has become essential in the management of hepatitis C virus (HCV) infection in order to diagnose infection, guide treatment decisions and assess the virological response to antiviral therapy. Virological tools include serological assays for anti-HCV antibody detection and serological determination of the HCV genotype, and molecular assays that detect and quantify HCV RNA and determine the HCV genotype. Anti-HCV antibody testing and HCV RNA testing are used to diagnose acute and chronic hepatitis C. Only patients with detectable HCV RNA should be considered for pegylated interferon alfa and ribavirin therapy and the HCV genotype should be systematically determined before treatment, as it determines the indication, the duration of treatment, the dose of ribavirin and the virological monitoring procedure. HCV RNA monitoring during therapy is used to tailor treatment duration in HCV genotype 1 infection, and molecular assays are used to assess the end-of-treatment and, most importantly the sustained virological response, i.e. the endpoint of therapy.
Keywords: Hepatitis C virus, serological and virological tests, HCV RNA
1. INTRODUCTION
Virological testing has become essential in the management of hepatitis C virus (HCV) infection in order to diagnose infection, and most importantly guide treatment decisions and assess the virological response to antiviral therapy.
2. VIROLOGICAL TOOLS
Serological assays
Anti-HCV antibody detection
The detection of anti-HCV antibodies in plasma or serum is based on the use of third-generation EIAs, that detect mixtures of antibodies directed against various HCV epitopes. Recombinant antigens are used to capture circulating anti-HCV antibodies onto the wells of microtiter plates, microbeads, or specific holders adapted to closed automated devices. The presence of anti-HCV antibodies is revealed by anti-antibodies labeled with an enzyme that catalyzes the transformation of a substrate into a colored compound. The optical density (OD) ratio of the reaction (sample OD/internal control OD) is proportional to the amount of antibodies in the serum or plasma sample [ 1 ]. The specificity of third-generation EIAs for anti-HCV is greater than 99% [ 2 ]. Their sensitivity is more difficult to determine, given the lack of a gold standard method, but it is excellent in HCV-infected immunocompetent patients. EIAs can be fully automated and are well adapted to large volume testing. Immunoblot tests are nowadays clinically obsolete given the good performance of third-generation anti-HCV EIAs [ 3 ].
Serological determination of the HCV genotype
The HCV genotype can be determined by seeking for antibodies directed to genotype-specific HCV epitopes with a competitive EIA. The currently available assay (Murex HCV serotyping 1-6 HC02, Abbott Laboratories, North Chicago, Illinois) identifies the type (1 to 6), but does not discriminate among the subtypes, and provides interpretable results in approximately 90% of chronically infected immunocompetent patients [ 4 ]. Mixed serological reactivities can be observed that could be related to mixed infection although cross-reactivity or recovery from one genotype infection and persistence of viremia with another genotype cannot be ruled out.
Detection and quantification of HCV RNA
Qualitative, non-quantitative HCV RNA detection
Qualitative detection assays are based on the principle of target amplification using either “classic” polymerase chain reaction (PCR), “real-time” PCR or TMA [ 5 ]. HCV RNA is extracted and reverse transcribed into a double stranded complementary DNA (cDNA), which is subsequently processed into a cyclic enzymatic reaction leading to the generation of a large number of detectable copies. Double-stranded DNA copies of HCV genome are synthesized in PCR-based assays, whereas single-stranded RNA copies are generated in TMA. Detection of amplified products is achieved by hybridizing the produced amplicons onto specific probes after the reaction in “classic” PCR or TMA techniques [ 5 ]. In “real-time” PCR, each round of amplification leads to the emission of a fluorescent signal and the number of signals per cycle is proportional to the amount of HCV RNA in the starting sample [ 5-7 ]. Qualitative detection assays must detect 50 HCV RNA IU/ml or less, and have equal sensitivity for the detection of all HCV genotypes. The lower limit of detection of the qualitative, non quantitative reverse-transcriptase PCR-based assay Amplicor® HCV v2.0, or of its semi-automated version Cobas® Amplicor® HCV v2.0 (Roche Molecular Systems, Pleasanton, California) is 50 IU/ml, whereas that of the TMA-based assay Versant® HCV RNA Qualitative Assay (Bayer HealthCare) is 10 IU/ml (Table 1). Real-time PCR assays, which are also able to quantify HCV RNA, have lower limits of detection of the order of 5-30 IU/ml when they are used as purely qualitative, non-quantitative assays.
Characteristics of current HCV RNA assays. RT : reverse transcriptase, PCR : polymerase chain reaction, TMA : transcription-mediated amplification, bDNA : “branched DNA“, NA : not applicable. *for 0.2 ml or 0.5 ml of plasma analyzed, respectively
| Assay | Manufacturer | Technique | Lower limit of detection (qualitative assay) | Dynamic range of quantification (quantitative assay) |
|---|---|---|---|---|
| Amplicor® HCV v2.0 | Roche Molecular Systems | Manual RT-PCR | 50 IU/ml | NA |
| Cobas® Amplicor® HCV v2.0 | Roche Molecular Systems | Semi-automated RT-PCR | 50 IU/ml | NA |
| Versant® HCV RNA Qualitative Assay | Bayer HealthCare | Manual TMA | 10 IU/ml | NA |
| Amplicor HCV Monitor® v2.0 | Roche Molecular Systems | Manual RT-PCR | 600 IU/ml | 600-500,000 IU/ml |
| Cobas® Amplicor HCV Monitor v2.0 | Roche Molecular Systems | Semi-automated RT-PCR | 600 IU/ml | 600-500,000 IU/ml |
| LCx HCV RNA Quantitative Assay | Abbott Diagnostic | Semi-automated RT-PCR | 25 IU/ml | 25-2,630,000 IU/ml |
| Versant® HCV RNA 3.0 Assay | Bayer HealthCare | Semi-automated bDNA | 615 IU/ml | 615-7,700,000 IU/ml |
| Cobas® TaqMan HCV Test | Roche Molecular Systems | Semi-automated real-time PCR | 15 IU/ml | 43-69,000,000 IU/ml |
| Abbott RealTime | Abbott Diagnostic | Semi-automated real-time PCR | 30 IU/ml or 12 IU/ml* | 12-100,000,000 IU/ml |
HCV RNA quantification
HCV RNA can be quantified by means of target amplification techniques (competitive PCR or real-time PCR) or signal amplification techniques (branched DNA (bDNA) assay) [ 5 ]. Five standardized assays are commercially available. Two of them are based on competitive PCR : Amplicor HCV Monitor® v2.0 and its semi-automated version Cobas® Amplicor HCV Monitor® v2.0 (Roche Molecular Systems), and LCx® HCV RNA Quantitative Assay (Abbott Diagnostic); one is based on bDNA technology, Versant® HCV RNA 3.0 Assay (Bayer Healthcare) ; and two are based on real-time PCR amplification, Cobas® TaqMan HCV Test, which can be coupled with automated extraction in Cobas Ampliprep® (Roche Molecular Systems), and Abbott RealTime™ HCV assay (Abbott Diagnostics), which uses the Abbott m2000 system and can also be coupled with an automated extraction procedure. Table 1 shows the respective dynamic ranges of quantification of the currently available assays, i.e. the HCV RNA intervals within which quantification is accurate in the corresponding assay. HCV RNA levels falling above the upper limit of quantification of the assay are underestimated and the samples must be retested after 1/10 to 1/100 dilution in order to achieve accurate quantification. The most promising approach for the future is fully automated real-time PCR assays, which are faster, more sensitive than classical target amplification techniques and are not prone to carryover contamination.
Molecular determination of the HCV genotype (genotyping)
The reference method for HCV genotype determination is direct sequencing of the NS5B or E1 regions of HCV genome by means of “in-house” techniques, followed by sequence alignment with prototype sequences and phylogenetic analysis [ 8, 9 ]. These techniques must be used in molecular epidemiology studies, where exact subtyping is needed. In clinical practice, HCV genotype can be determined by various commercial kits, using direct sequence analysis of the 5' noncoding region (Trugene® 5'NC HCV Genotyping Kit, Bayer HealthCare, Diagnostics Division, Tarrytown, New York) or reverse hybridization analysis using genotype-specific probes located in the 5' noncoding region (commercialized as INNO-LiPA HCV II, Innogenetics, Ghent, Belgium, or Versant® HCV Genotyping Assay, Bayer HealthCare) [10-13 ]. Mistyping is rare with these techniques, but mis-subtyping may occur in 10 to 25% of cases, related to the studied region (5' noncoding region) rather than the technique used. These errors have no clinical consequences, because only the type is used for therapeutic decision-making. An assay based on direct sequencing of the NS5B region is currently in development (Trugene® NS5B HCV Genotyping Kit, Bayer HealthCare).
3. DIAGNOSIS OF HCV INFECTION
Acute hepatitis C
Patients with a suspicion of acute hepatitis C should be tested for both anti-HCV antibodies by EIA and HCV RNA with a sensitive technique, i.e. an HCV RNA assay with a lower limit of detection of 50 IU/ml or less [ 1 ]. Four marker profiles can be observed according to the presence or absence of either marker. The presence of HCV RNA in the absence of anti-HCV antibodies is strongly indicative of acute HCV infection, which will be confirmed by seroconversion (i.e. the appearance of anti-HCV antibodies) a few days to weeks later. Acutely infected patients can also have both HCV RNA and anti-HCV antibodies at the time of diagnosis. It is difficult, in this case, to distinguish acute hepatitis C from an acute exacerbation of chronic hepatitis C or an acute hepatitis of another cause in a patient with chronic hepatitis C. Acute hepatitis C is very unlikely if both anti-HCV antibodies and HCV RNA are absent. It is also unlikely if anti-HCV antibodies are present without HCV RNA. These patients should however be retested after a few weeks because HCV RNA can be temporarily undetectable, due to transient, partial control of viral replication by the immune response before replication escapes and chronic infection establishes [14 ]. Apart from such cases, the presence of anti-HCV antibodies in the absence of HCV RNA is generally seen in patients who have recovered from a past HCV infection. Nevertheless, this pattern cannot be differentiated from a false positive EIA result, the exact prevalence of which is unknown.
Chronic hepatitis C
In patients with clinical or biological signs of chronic liver disease, chronic hepatitis C is certain when both anti-HCV antibodies and HCV RNA (sought for with a sensitive technique, detecting 50 IU/ml or less) are present [ 3, 15 ]. Detectable HCV replication in the absence of anti-HCV antibodies is exceptional with the current third-generation EIAs, almost exclusively observed in profoundly immunodepressed patients, hemodialysis patients or agammaglobulinemic subjects [16, 17 ].
In patients who have no indication for therapy or have a contra-indication to the use of antiviral drugs, virological tests have no prognostic value. Indeed, neither anti-HCV antibodies nor the HCV RNA load correlate with the severity of liver inflammation or fibrosis nor with their progression. Thus, they cannot be used to predict the natural course of infection or the onset of extrahepatic manifestations. In untreated patients, the severity of liver inflammation and fibrosis must be evaluated every three to five years by means of a liver biopsy or non-invasive serological or ultrasound-based testing [18 ].
4. MANAGEMENT OF ANTIVIRAL THERAPY
The current standard treatment for chronic hepatitis C is the combination of pegylated interferon (IFN) alfa and ribavirin [18 ]. The efficacy endpoint of hepatitis C treatment is the “sustained virological response” (SVR), defined by the absence of detectable HCV RNA in serum as assessed by an HCV RNA assay with a lower limit of detection of 50 IU/ml or less 24 weeks after the end of treatment [18 ].
Initiation of therapy
Only patients with detectable HCV RNA should be considered for pegylated IFN alfa and ribavirin combination therapy [18 ]. The decision to treat patients with chronic hepatitis C depends on multiple parameters, including a precise assessment of the severity of liver disease and of its foreseeable outcome, the presence of absolute or relative contra-indications to therapy, and the patient's willingness to be treated.
The HCV genotype should be systematically determined before treatment, as it determines the indication, the duration of treatment, the dose of ribavirin and the virological monitoring procedure [19 ].
HCV genotype 1
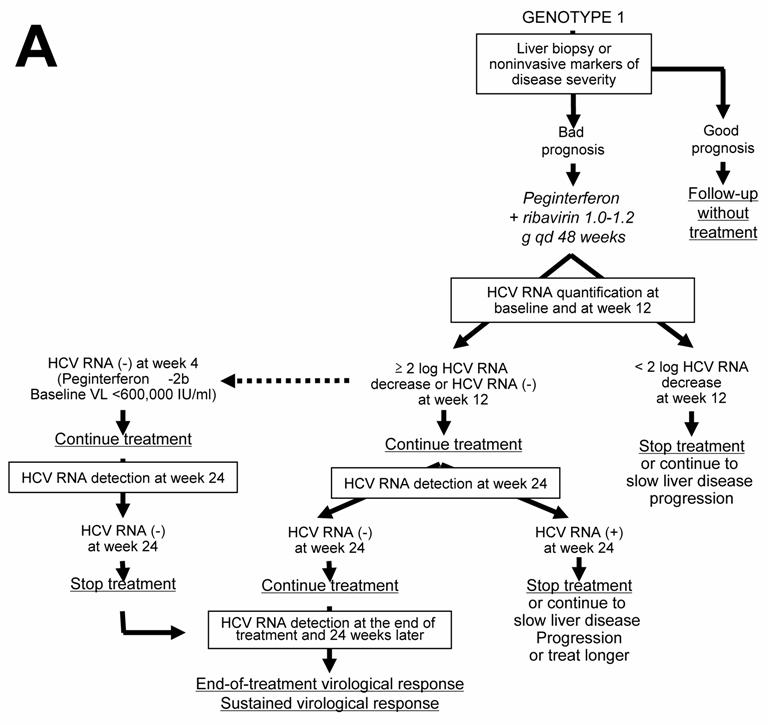
Given the likelihood of a sustained virological response, of the order of 40% to 50%, a precise assessment of liver disease prognosis by means of a liver biopsy or a non-invasive method based on serological markers of fibrosis or ultrasound-based testing [20, 21 ] must be performed in order to help with the treatment decision (Figure 1A). It is recommended not to treat patients with mild lesions and to re-assess their liver disease after 3 to 5 years. The patients with inflammation and/or fibrosis (Metavir score A ≥ 2 and/or F ≥ 2) have an indication for therapy [18 ].
The approved dose of pegylated IFN alfa-2a is 180 µg per week, independent of body weight, whereas that of pegylated IFN alfa-2b is weight-adjusted at 1.5 µg/kg per week, identical for all HCV genotypes. Patients infected with HCV genotype 1 should receive a high dose of ribavirin, i.e. 1,000 to 1,200 mg daily, based on body weight less than or greater than 75 kg (it has been recently suggested that the heaviest patients could even benefit from a higher ribavirin dose, up to 1,600 mg daily) and they theoretically require 48 weeks of treatment (Figure 1A) [18 ]. However, monitoring of HCV RNA load decrease during therapy is recommended in order to avoid treating for 48 weeks patients with no likelihood of an SVR [22, 23 ]. In this purpose, HCV RNA quantification should be performed at baseline and after 12 weeks of treatment (Figure 1A) [18 ]. Both measures must be performed with the same technique in order to ensure comparability of the results at the two time points. Treatment must be continued when there is a 2-log drop in HCV RNA level, i.e. when baseline HCV RNA level is divided by 100 or more, or when HCV RNA is undetectable at week 12 [18 ]. In these patients, it is recommended to assess the presence of HCV RNA with a sensitive technique (lower limit of detection : 50 IU/ml or less) at week 24. If HCV RNA is undetectable at week 24, treatment must be continued until week 48, with a high likelihood of an SVR. It was recently suggested that 24 weeks of therapy might be sufficient for patients with a baseline viral load below 600,000 IU/ml in whom pegylated IFN alfa-2b-based treatment yields a 2-log decline at week 12 and undetectable HCV RNA at week 24 [24 ]. In contrast, if HCV RNA is still detectable at week 24, the likelihood of an SVR is virtually nil and treatment can be stopped or continued with the only aim to slow liver disease progression in patients with a severe prognosis, without any hope to eradicate infection (Figure 1A) [18, 22 ]. Ongoing trials are studying whether a prolonged antiviral treatment or maintenance therapy with pegylated IFN alfa monotherapy could be beneficial in the latter patients. When treatment is continued until week 48, the end-of-treatment and sustained virological responses should be assessed by means of a sensitive HCV RNA assay, with a lower limit of detection of 50 IU/ml or less [18 ]. HCV RNA detection at the end of therapy is highly predictive of a post-treatment relapse, whereas the absence of HCV RNA at the end of treatment indicates a virological response. These patients must be retested for HCV RNA with a sensitive method 24 weeks later in order to assess the SVR, i.e. the endpoint of therapy [1, 18 ]. HCV infection appears to be definitively cured in the vast majority of sustained virological responders.
The lack of a 12-week virological response (no change or an HCV RNA decrease of less than 2 logs at week 12) is associated with a virtually nil probability of a subsequent sustained virological response [22, 23 ]. Treatment can thus be stopped at week 12 in these patients, or continued to slow liver disease progression without clearing the virus (Figure 1A). The benefits of maintenance therapy on the outcome of HCV-associated liver disease are currently under investigation. This “stopping rule”, based on monitoring of HCV RNA load reduction at week 12, was recently shown to also apply to patients co-infected with HCV and human immunodeficiency virus [25-27 ].
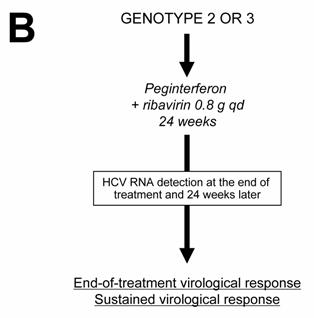
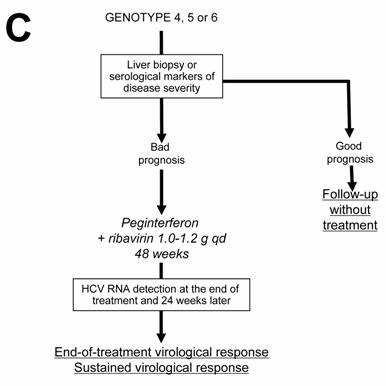
Current algorithms for the use of HCV virological tools in the treatment of chronic hepatitis C, according to the HCV genotype: genotype 1 (A), genotypes 2 and 3 (B), and genotypes 4, 5 and 6 (C)
HCV genotypes 2 and 3
Patients infected with HCV genotypes 2 or 3 have a 70%-80% likelihood of an SVR with a low dose of ribavirin and only 24 weeks of treatment [19, 23, 28 ]. Thus, in the absence of contra-indications, these patients should be treated regardless of the severity of their liver disease and they do not need a liver biopsy of noninvasive assessment of liver fibrosis (Figure 1B). The recommended dose of pegylated IFN alfa-2a or alfa-2b is the same as for HCV genotype 1, i.e. 180 μg/week and 1.5 μg/kg/week, respectively. The fixed recommended dose of ribavirin is 800 mg per day (Figure 1B) [18 ]. It is possible that even lower doses of ribavirin and/or shorter duration of treatment could be sufficient to achieve an SVR in certain subgroups of patients with genotype 2 or 3 infection, such as those with a low baseline viral load and no extensive fibrosis or cirrhosis, as suggested by recent preliminary data [29 ]. One should be careful in patients who combine several baseline parameters of non-response, such as extensive fibrosis, an old age and a male gender, who might need 48 weeks of therapy to clear infection.
No monitoring of HCV RNA level changes during therapy is recommended in the patients with genotype 2 or 3 infection, because the vast majority of them become HCV RNA-negative early during treatment. Like in HCV genotype 1-infected patients, the virological response must be assessed by means of a sensitive HCV RNA assay at the end of therapy and 24 weeks later in order to determine whether the virological response is sustained (Figure 1B) [1, 18 ].
HCV genotypes 4, 5 and 6
In the absence of any clinical trial including a sufficient number of patients, the likelihood of an SVR and the optimal treatment schedule remain unknown for the patients infected with HCV genotypes 4, 5 or 6. It is thus recommended to treat them like those infected with HCV genotype 1, i.e. with pegylated IFN alfa at the usual dose, combined with a high dose of ribavirin (1000-1200 mg per day, according to body weight less or greater than 75 kg) (Figure 1C). In the absence of published data, no stopping rules have been defined and it is recommended to treat these patients for a total of 48 weeks. The virological response must be assessed by means of a sensitive HCV RNA assay (lower limit of detection of 50 IU/ml or less) at the end of therapy and 24 weeks later [1, 18 ].
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Pawlotsky JM. Use and interpretation of virological tests for hepatitis C. Hepatology. 2002 ;36(Suppl 1):S65-73
2. Colin C. et al. Sensitivity and specificity of third-generation hepatitis C virus antibody detection assays: an analysis of the literature. J Viral Hepat. 2001 ;8:87-95
3. Pawlotsky JM. et al. What strategy should be used for diagnosis of hepatitis C virus infection in clinical laboratories? Hepatology. 1998 ;27:1700-2
4. Pawlotsky JM. et al. Serological determination of hepatitis C virus genotype: comparison with a standardized genotyping assay. J Clin Microbiol. 1997 ;35:1734-9
5. Pawlotsky JM. Molecular diagnosis of viral hepatitis. Gastroenterology. 2002 ;122:1554-68
6. Martell M. et al. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999 ;37:327-32
7. Komurian-Pradel F. et al. Quantitation of HCV RNA using real-time PCR and fluorimetry. J Virol Methods. 2001 ;95:111-9
8. Simmonds P. Viral heterogeneity of the hepatitis C virus. J Hepatol. 1999 ;31(Suppl 1):54-60
9. Simmonds P. et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005 ;42:962-73
10. Germer JJ. et al. Automated sample preparation for the Trugene HIV-1 genotyping kit using the MagNA pure LC instrument. Diagn Microbiol Infect Dis. 2004 ;49:59-61
11. Stuyver L. et al. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996 ;34:2259-66
12. Stuyver L. et al. Hepatitis C virus genotyping by means of 5'-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 1995 ;38:137-57
13. Zheng X. et al. Direct comparison of hepatitis C virus genotypes tested by INNO-LiPA HCV II and TRUGENE HCV genotyping methods. J Clin Virol. 2003 ;28:214-6
14. Lavillette D. et al. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005 ;79:6023-34
15. EASL International Consensus Conference on Hepatitis C. Paris, 26-28, February 1999, Consensus Statement. European Association for the Study of the Liver. J Hepatol. 1999 ;30:956-61
16. Lok AS. et al. Antibody response to core, envelope and nonstructural hepatitis C virus antigens: comparison of immunocompetent and immunosuppressed patients. Hepatology. 1993 ;18:497-502
17. Thio CL. et al. Screening for hepatitis C virus in human immunodeficiency virus-infected individuals. J Clin Microbiol. 2000 ;38:575-7
18. NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens State Sci Statements. 2002 ;19:1-46
19. Hadziyannis SJ. et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004 ;140:346-55
20. Castera L. et al. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology. 2005 ;128:343-50
21. Poynard T. et al. FibroTest-FibroSURE: towards a universal biomarker of liver fibrosis? Expert Rev Mol Diagn. 2005 ;5:15-21
22. Davis GL. et al. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology. 2003 ;38:645-52
23. Fried MW. et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002 ;347:975-82
24. Zeuzem S. et al. Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J Hepatol. 2006 ;44:97-103
25. Chung RT. et al. Peginterferon Alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004 ;351:451-9
26. Pawlotsky JM. Treating hepatitis C in "difficult-to-treat" patients. N Engl J Med. 2004 ;351:422-3
27. Torriani FJ. et al. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004 ;351:438-50
28. Manns MP. et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001 ;358:958-65
29. Dalgard O. et al. Treatment with pegylated interferon and ribavarin in HCV infection with genotype 2 or 3 for 14 weeks: a pilot study. Hepatology. 2004 ;40:1260-5
Author biography
Stéphane Chevaliez, PharmD, PhD, is Assistant Professor at the University of Paris XII and a member of the Department of Virology and of INSERM Unit U635 at the Henri Mondor University Hospital in Créteil, France. He focuses on teaching, diagnosis and research in hepatitis viruses. He earned his medical degree and a Thesis in molecular virology from the University of Paris and Pasteur Institute, France.
Jean-Michel Pawlotsky, MD, PhD, is Professor of Medicine at the University of Paris XII and the Director of the Department of Virology, of INSERM Unit U635 and of the French National Reference Center on Viral Hepatitis B, C and delta at the Henri Mondor University Hospital in Créteil, France. He focuses on teaching, diagnosis and research in virology, primarily hepatitis C and hepatitis viruses. He earned his medical degree and a Thesis in molecular virology from the University of Paris, France. In addition, Dr Pawlotsky is a graduate in virology from the Pasteur Institute in Paris and microbiology from the University of Paris. He is active in numerous professional societies, currently acting as the Scientific Secretary of the European Association for the Study of the Liver (EASL), and he is a member of the Scientific College of the French National Agency for AIDS and Viral Hepatitis Research (ANRS). Dr Pawlotsky is currently an Associate Editor of Hepatology, the official journal of the American Association for the Study of Liver Diseases (AASLD), and of Current Hepatitis Reports, and a member of the Editorial Board of the Journal of Hepatology and of the Journal of Virological Methods. Dr Pawlotsky's noted career contributions include the publication of over 300 articles and book chapters in his areas of expertise.
![]() Corresponding address:
Corresponding address:
Professor Jean-Michel PAWLOTSKY, M.D., Ph.D., Department of Virology, Hôpital Henri Mondor, 51 avenue du Maréchal de Lattre de Tassigny, 94010 Créteil, France. Tel : +33-1-4981-2827, Fax : +33-1-4981-4831. E-mail : jean-michel.pawlotskyaphp.fr

 Global reach, higher impact
Global reach, higher impact